import GODE_ebayesThis tutorial is about GODE: Graph fourier transform based Outlier Detection using Emprical Bayesian thresholding paper.
0. Import
import numpy as np
import pandas as pdfrom pygsp import graphs, filters, plotting, utils
import pickle1. Linear
1.1. Data
Data description
- Graph \({\cal G}=({\cal V},E,{\bf W})\)
- Vertex set \({\cal V}=\{1,2,\dots,n\}=\{ v_1,v_2,\dots,v_N\}\)
- \(N=1000\)
- \(W_{ij} = \begin{cases} 1, & \text{ if } j-i = 1 \\ 0, & \text{ otherwise}\end{cases}\)
- Graph signal \(y:{\cal V} \to \mathbb{R}\)
\[y_i=\frac{10}{N}v_i+\eta_i+\epsilon_i,\]
- \(\eta_i = \begin{cases} U(5,7) & \text{ with probability 0.025}\\ U(-7,-5) & \text{ with probability 0.025} \\ 0 & \text{ otherwise } \end{cases}\)
- \(\epsilon_i \sim N(0,1)\)
Generate Dataset
np.random.seed(6)
n = 1000
eta_sparsity = 0.05
epsilon = np.around(np.random.normal(size=n),15)
signal = np.random.choice(np.concatenate((np.random.uniform(-7, -5, round(n*eta_sparsity/2)).round(15), np.random.uniform(5, 7, round(n*eta_sparsity/2)).round(15), np.repeat(0, n - round(n*eta_sparsity)))), n)
eta = signal + epsilon
outlier_true_linear= signal.copy()
outlier_true_linear = list(map(lambda x: 1 if x!=0 else 0,outlier_true_linear))
x_1 = np.linspace(0,2,n)
y1_1 = 5 * x_1
y_1 = y1_1 + eta # eta = signal + epsilon
_df=pd.DataFrame({'x':x_1, 'y':y_1})
index_of_trueoutlier_bool = signal!=01.2. GODE
Lin = GODE_ebayes.Linear(_df)Lin.fit(sd=20)outlier_old_linear, outlier_linear, outlier_index_linear = GODE_ebayes.GODE_Anomalous(Lin(_df),contamination=0.05)1.3. Plot
GODE_ebayes.Linear_plot(Lin(_df),index_of_trueoutlier_bool, outlier_index_linear)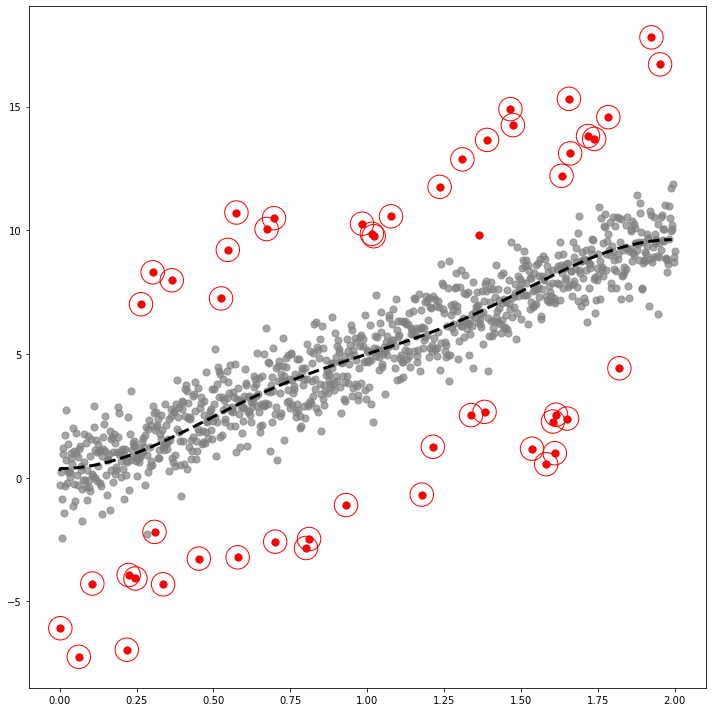
how to access data
Lin(_df)['x'][:10]array([0. , 0.002002 , 0.004004 , 0.00600601, 0.00800801,
0.01001001, 0.01201201, 0.01401401, 0.01601602, 0.01801802])Lin(_df)['y'][:10]array([-0.31178367, -6.08358567, 0.23784081, -0.86906177, -2.44674061,
0.96330157, 1.18712379, -1.44402316, 1.71937116, -0.33980351])Lin(_df)['yhat'][:10]array([0.26224717, 0.37091714, 0.37104804, 0.3712662 , 0.3715716 ,
0.37196424, 0.37244408, 0.37301111, 0.3736653 , 0.37440661])1.4. Confusion matrix
Conf_linear = GODE_ebayes.Conf_matrx(outlier_true_linear,outlier_linear)Conf_linear.conf('GODE')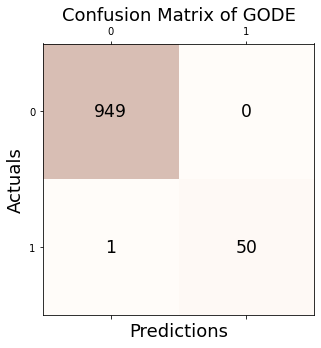
Accuracy: 0.999
Precision: 1.000
Recall: 0.980
F1 Score: 0.990Conf_linear(){'Accuracy': 0.999,
'Precision': 1.0,
'Recall': 0.9803921568627451,
'F1 Score': 0.99009900990099}2. Orbit
2.1. Data
Data description
- Graph \({\cal G}=({\cal V},E,{\bf W})\)
- \(N=1000\)
- \({\boldsymbol v}_i=(r_i \cos(\theta_i), r_i \sin(\theta_i))\)
- \(r_i= 5 + \cos(\frac{12\pi (i - 1)}{n - 1})\)
- \(\theta_i= -\pi + \frac{{\pi(N-2)(i - 1)}}{N(N - 1)}\)
- graph signal \(y:{\cal V} \to \mathbb{R}\)
\[y_i = 10 \cdot \sin\left(\frac{{6\pi \cdot (i - 1)}}{{N - 1}}\right)+\epsilon_i+\eta_i\]
- \(\eta_i = \begin{cases} U(1,4) & \text{ with probability 0.025}\\ U(-4,-1) & \text{ with probability 0.025} \\ 0 & \text{ otherwise } \end{cases}\)
- \(\epsilon_i \sim N(0,1)\)
- \({\bf W} = \begin{cases} \exp\left(-\frac{|{\boldsymbol v}_i -{\boldsymbol v}_j|^2}{2\theta^2}\right), & \text{ if } |{\boldsymbol v}_i - {\boldsymbol v}_j| \le \kappa \\ 0, & \text{ otherwise}\end{cases}\)
- \(\theta=6.45\) and \(\kappa=1.21\)
Generate Dataset
n = 1000
eta_sparsity = 0.05
random_seed=77
np.random.seed(777)
epsilon = np.around(np.random.normal(size=n),15)
signal = np.random.choice(np.concatenate((np.random.uniform(-4, -1, round(n * eta_sparsity / 2)).round(15), np.random.uniform(1, 4, round(n * eta_sparsity / 2)).round(15), np.repeat(0, n - round(n * eta_sparsity)))), n)
eta = signal + epsilon
pi=np.pi
ang=np.linspace(-pi,pi-2*pi/n,n)
r=5+np.cos(np.linspace(0,12*pi,n))
vx=r*np.cos(ang)
vy=r*np.sin(ang)
f1=10*np.sin(np.linspace(0,6*pi,n))
f = f1 + eta
_df = pd.DataFrame({'x' : vx, 'y' : vy, 'f' : f,'f1':f1})
outlier_true_orbit = signal.copy()
outlier_true_orbit = list(map(lambda x: 1 if x!=0 else 0,outlier_true_orbit))
index_of_trueoutlier_bool = signal!=02.2. GODE
Or = GODE_ebayes.Orbit(_df)Or.fit(sd=15,method = 'Euclidean',kappa=1.21)100%|██████████| 1000/1000 [00:01<00:00, 683.67it/s]outlier_old_orbit, outlier_orbit, outlier_index_orbit = GODE_ebayes.GODE_Anomalous(Or(_df),contamination=0.05)2.3. Plot
GODE_ebayes.Orbit_plot(Or(_df),index_of_trueoutlier_bool,outlier_index_orbit)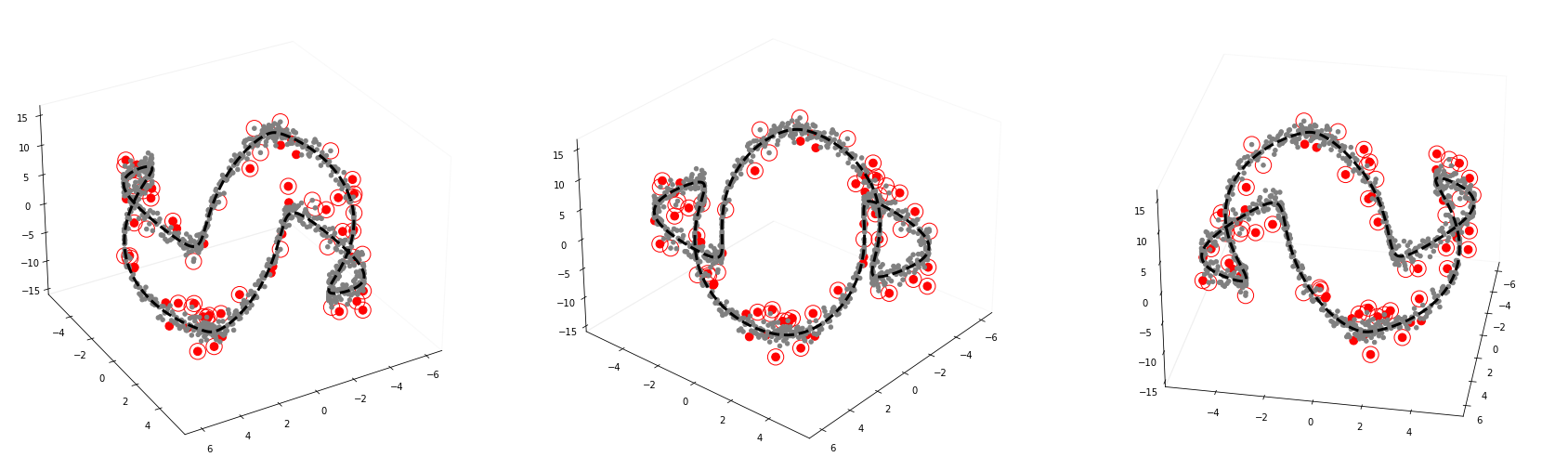
how to access data
Or(_df)['x'][:5]array([-6. , -5.99916963, -5.9966797 , -5.99253378, -5.98673778])Or(_df)['y'][:5]array([-7.34788079e-16, -3.76943905e-02, -7.53604665e-02, -1.12969981e-01,
-1.50494820e-01])Or(_df)['f1'][:5]array([0. , 0.18867305, 0.37727893, 0.56575049, 0.75402065])Or(_df)['f'][:5]array([-0.46820879, -0.63415181, 0.31189882, -0.14761143, 1.66037153])Or(_df)['fhat'][:5]array([-0.12358846, 0.28544397, 0.69162952, 0.75432307, 0.83443358])2.4. Confusion matrix
Conf_orbit = GODE_ebayes.Conf_matrx(outlier_true_orbit,outlier_orbit)Conf_orbit.conf('GODE')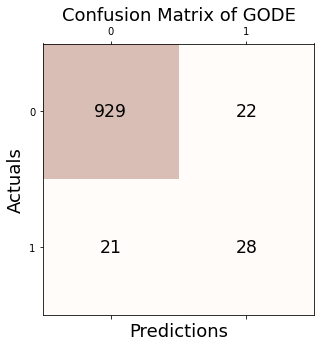
Accuracy: 0.957
Precision: 0.560
Recall: 0.571
F1 Score: 0.566Conf_orbit(){'Accuracy': 0.957,
'Precision': 0.56,
'Recall': 0.5714285714285714,
'F1 Score': 0.5656565656565656}3. Bunny
3.1. Data
Data description
- Stanford bunny data
- \(n=2503\)
Generate Dataset
eta_sparsity = 0.05
random_seed=77
n = 2503
with open("Bunny.pkl", "rb") as file:
loaded_obj = pickle.load(file)
_df = pd.DataFrame({'x':loaded_obj['x'],'y':loaded_obj['y'],'z':loaded_obj['z'],'f':loaded_obj['f']+loaded_obj['noise'],'f1':loaded_obj['f'],'noise':loaded_obj['noise']})
outlier_true_bunny = loaded_obj['unif'].copy()
outlier_true_bunny = list(map(lambda x: 1 if x !=0 else 0,outlier_true_bunny))
index_of_trueoutlier_bool_bunny = loaded_obj['unif']!=0
_W = loaded_obj['W'].copy()3.2. GODE
bu = GODE_ebayes.BUNNY(_df,_W)bu.fit(sd=20)outlier_old_bunny, outlier_bunny, outlier_index_bunny = GODE_ebayes.GODE_Anomalous(bu(_df),contamination=eta_sparsity)3.3. Plot
GODE_ebayes.Bunny_plot(bu(_df),index_of_trueoutlier_bool_bunny,outlier_index_bunny)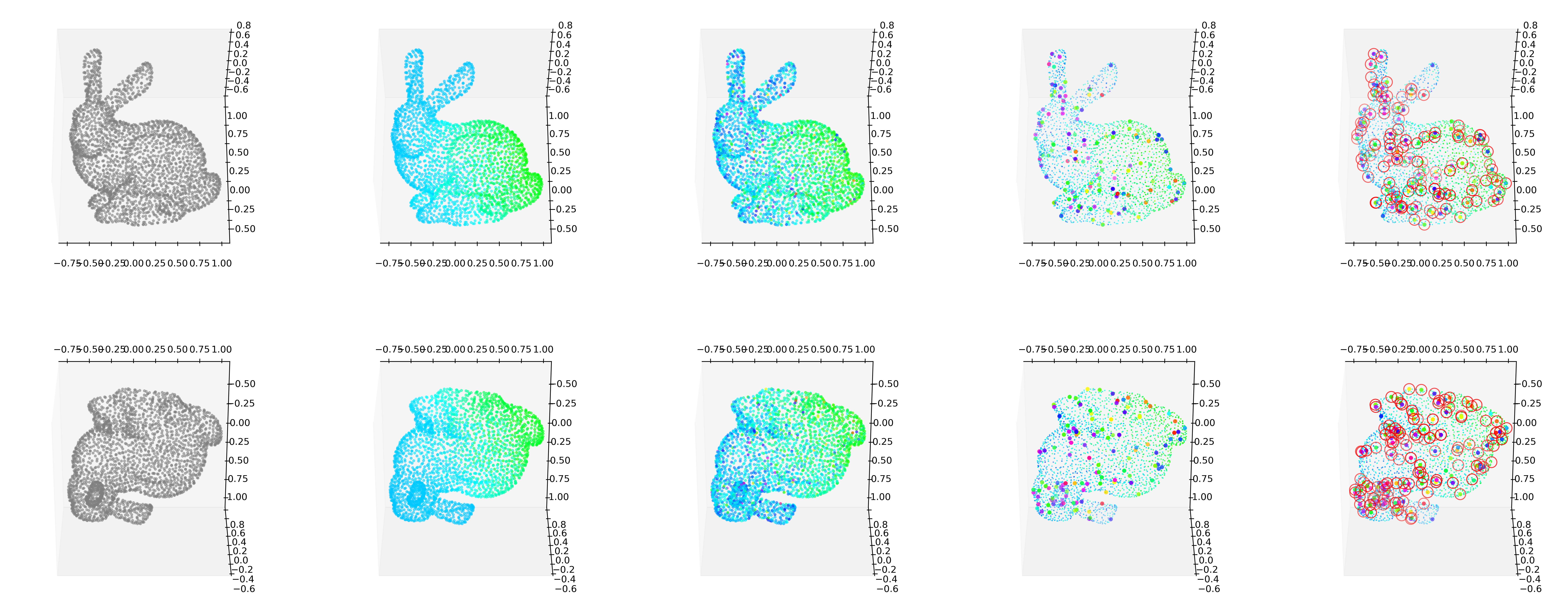
how to access data
bu(_df){'x': array([ 0.26815193, -0.58456893, -0.02730755, ..., 0.15397547,
-0.45056488, -0.29405249]),
'y': array([ 0.39314334, 0.63468595, 0.33280949, ..., 0.80205526,
0.6207154 , -0.40187451]),
'z': array([-0.13834514, -0.22438843, 0.08658215, ..., 0.33698514,
0.58353051, -0.08647485]),
'f1': array([-1.54422488, -0.03596483, -0.93972715, ..., -0.01924028,
-0.02470869, -0.26266752]),
'f': array([-1.63569131, 0.49423926, -1.04026277, ..., -1.0694093 ,
-0.24395499, 0.41729667]),
'fhat': array([-1.66060553, 0.09516849, -1.11731239, ..., 0.23695946,
0.16766298, -0.11652469])}3.4. Confusion matrix
Conf_bunny = GODE_ebayes.Conf_matrx(outlier_true_bunny,outlier_bunny)Conf_bunny.conf('GODE')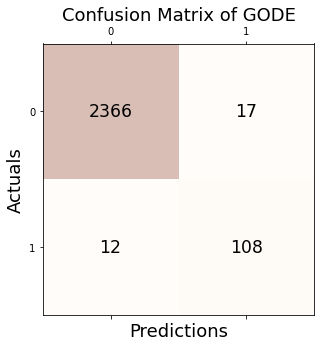
Accuracy: 0.988
Precision: 0.864
Recall: 0.900
F1 Score: 0.882Conf_bunny(){'Accuracy': 0.9884139033160207,
'Precision': 0.864,
'Recall': 0.9,
'F1 Score': 0.8816326530612244}4. Earthquake
4.1. Data
Data description
USGS data from \(2010\) to \(2014\)
Graph \({\cal G}=({\cal V},E,{\bf W})\)
Vertex set \({\cal V}= \{v_1, v_2, \ldots, v_n\}\)
\(v:= \tt{( Latitude}\), \(\tt{Longitude)}\)
\(N=10762\)
\({\bf W_{ij}} = \begin{cases} \exp(-\frac{\rho(i,j)}{2 \theta^2}), & \text{ if } |\rho(i,j)| \le \kappa \\ 0, & \text{ otherwise}\end{cases}\)
\(\rho(i,j)=hs({\boldsymbol v}_i, {\boldsymbol v}_j)\)
\(\theta = 8810.87\) and \(\kappa = 2500\)
graph signal \(y:{\cal V} \to \mathbb{R}\)
_df = pd.read_csv('./earthquake_tutorial.csv')_df = _df.assign(Year=list(map(lambda x: x.split('-')[0], _df.time))).rename(columns={'latitude' : 'x', 'longitude' : 'y', 'mag': 'f'}).iloc[:,1:]
_df.Year = _df.Year.astype(np.float64)4.2. GODE
Er = GODE_ebayes.Earthquake(_df.query("2010 <= Year < 2011"))Er.fit(sd=20, method = 'Haversine',kappa=2500)100%|██████████| 4790/4790 [00:30<00:00, 159.66it/s]outlier_old_earthquake, outlier_earthquake, outlier_index_earthquake = GODE_ebayes.GODE_Anomalous(Er(_df.query("2010 <= Year < 2011")),contamination=0.05)4.3. Plot
GODE_ebayes.Earthquake_plot(Er(_df.query("2010 <= Year < 2011")),outlier_index_earthquake,lat_center=37.7749, lon_center=-122.4194,fThresh=7,adjzoom=5,adjmarkersize = 40)how to access data
Er(_df.query("2010 <= Year < 2011")){'x': array([ 0.663, -19.209, -31.83 , ..., 40.726, 30.646, 26.29 ]),
'y': array([ -26.045, 167.902, -178.135, ..., 51.925, 83.791, 99.866]),
'f': array([5.5, 5.1, 5. , ..., 5. , 5.2, 5. ]),
'fhat': array([5.55678619, 5.05719136, 5.02115785, ..., 5.49869582, 5.45532648,
5.42616814])}