import itstgcn
import torch
import pandas as pd
import numpy as np
import randomData management for ITSTGCN
- 순환형 구조를 가진 모델(Models with recurrent structures):
- GConvGRU: 그래프 순환 유닛 (Graph Convolutional Gated Recurrent Unit)
- GConvLSTM: 그래프 순환 LSTM (Graph Convolutional Long Short-Term Memory)
- GCLSTM: 그래프 순환 LSTM (Graph Convolutional LSTM)
- DCRNN: 이중 그래프 순환 신경망 (Dual Graph Convolutional Recurrent Neural Network)
- 그래프 합성곱 모델(Graph convolution models):
- LRGCN: 로컬-전역 그래프 합성곱 신경망 (Local-Global Graph Convolutional Network)
- TGCN: 전이 그래프 합성곱 신경망 (Transition Graph Convolutional Network)
- EvolveGCNO: 진화형 그래프 합성곱 네트워크 (Evolutionary Graph Convolutional Network with Optimization)
- EvolveGCNH: 진화형 그래프 합성곱 네트워크 (Evolutionary Graph Convolutional Network with Hypernetwork)
- 동적 값 업데이트를 활용한 모델(Models utilizing dynamic value updates):
- DyGrEncoder: 동적 그래프 인코더 (Dynamic Graph Encoder)
논문에서 제안하는 방법은 GConvGRU (Chebyshev Graph Convolutional Gated Recurrent Unit Cell)와 GConvLSTM (Graph Convolutional Recurrent Network)이라는 두 가지 모델을 소개합니다. GConvGRU는 그래프 데이터에 대한 시간적인 의존성을 캡처하는데 사용되며, GConvLSTM은 그래프와 시퀀스 데이터를 동시에 처리하는데 활용됩니다.
GConvGRU는 Chebyshev 그래프 합성곱과 Gated Recurrent Unit (GRU)을 결합하여 그래프 데이터의 시간적 의존성을 모델링합니다. 이 모델은 그래프 내 노드들 간의 연결과 그래프의 구조를 고려하여 시계열 데이터를 예측하고 분석하는데 유용합니다.
“GC-LSTM: 그래프 합성곱 임베딩 LSTM을 이용한 동적 링크 예측”이라는 논문은 동적인 그래프에서 링크 예측 작업에 그래프 합성곱 네트워크(GCN)와 Long Short-Term Memory(LSTM) 셀을 결합한 새로운 모델을 제안합니다.
논문에서는 그래프 구조가 시간에 따라 변화하는 동적인 그래프에서 노드들 사이의 링크 존재 여부를 예측하는 문제를 다룹니다. 기존의 링크 예측 방법들은 동적인 그래프의 변화를 처리하는데 어려움이 있습니다.
제안하는 GC-LSTM 모델은 그래프 합성곱을 사용하여 노드의 특징을 임베딩하고 그래프 구조를 파악합니다. 동시에 LSTM 셀을 활용하여 시간적 의존성과 순차적 패턴을 모델링합니다.
GC-LSTM 모델은 동적인 그래프 데이터를 시간 순서대로 처리하여 변화하는 그래프 구조를 적절히 반영하고 미래의 링크를 정확하게 예측할 수 있습니다. 실험과 평가를 통해 GC-LSTM 모델이 동적인 링크 예측 작업에서 다른 최신 기법들보다 우수한 성능을 보여주며, 동적인 그래프 구조를 다루는데 유용한 접근 방법임을 입증합니다.
- GNAR stationary condition
data_dict = itstgcn.load_data('./data/fivenodes.pkl')
loader = itstgcn.DatasetLoader(data_dict)dataset = loader.get_dataset(lags=2)sum_result = 0
for i in range(5):
sum_result += np.sum(np.array(dataset.targets)[:,i])
print(sum_result)-28.048902156588262from torch_geometric_temporal.dataset import ChickenpoxDatasetLoader
loader1 = ChickenpoxDatasetLoader()dataset1 = loader1.get_dataset(lags=4)sum_result = 0
for i in range(20):
sum_result += np.sum(np.array(dataset1.targets)[:,i])
print(sum_result)9.553510148236436from torch_geometric_temporal.dataset import PedalMeDatasetLoader
loader2 = PedalMeDatasetLoader()dataset2 = loader2.get_dataset(lags=4)sum_result = 0
for i in range(15):
sum_result += np.sum(np.array(dataset2.targets)[:,i])
print(sum_result)-39.06545396690812from torch_geometric_temporal.dataset import WikiMathsDatasetLoader
loader3 = WikiMathsDatasetLoader()dataset3 = loader3.get_dataset(lags=8)sum_result = 0
for i in range(1068):
sum_result += np.sum(np.array(dataset3.targets)[:,i])
print(sum_result)-1543.6160620151331loader6 = itstgcn.load_data('./data/Windmillsmall.pkl')dataset6 = loader6.get_dataset(lags=8)sum_result = 0
for i in range(11):
sum_result += np.sum(np.array(dataset6.targets)[:,i])
print(sum_result)0.33864788949161806from torch_geometric_temporal.dataset import MontevideoBusDatasetLoader
loader10 = MontevideoBusDatasetLoader()dataset10 = loader10.get_dataset(lags=8)sum_result = 0
for i in range(675):
sum_result += np.sum(np.array(dataset10.targets)[:,i])
print(sum_result)1114.5722422676997Import
import pandas as pd
import matplotlib.pyplot as plt- GConvGRU
df_GConvGRU = pd.read_csv('./simulation_results/Real_simulation_reshape/Final_Simulation_GConvGRU.csv')
pedal_wiki_GSO_GConvGRU = pd.read_csv('./simulation_results/Real_simulation_reshape/Final_Simulation_GConvGRU_pedal_wiki_GSO.csv')df_GConvGRU['model']='GConvGRU'
pedal_wiki_GSO_GConvGRU['model']='GConvGRU'df_GConvGRU['dataset'].unique()array(['fivenodes', 'chickenpox', 'pedalme', 'wikimath', 'windmillsmall',
'monte'], dtype=object)- GConvLSTM
df_GConvLSTM = pd.read_csv('./simulation_results/Real_simulation_reshape/Final_Simulation_GConvLSTM.csv')
pedal_wiki_GSO_GConvLSTM = pd.read_csv('./simulation_results/Real_simulation_reshape/Final_Simulation_GConvLSTM_pedal_wiki_GSO.csv')df_GConvLSTM['model']='GConvLSTM'
pedal_wiki_GSO_GConvLSTM['model']='GConvLSTM'df_GConvLSTM['dataset'].unique()array(['fivenodes', 'chickenpox', 'pedalme', 'wikimath', 'windmillsmall',
'monte'], dtype=object)- GCLSTM
df_GCLSTM = pd.read_csv('./simulation_results/Real_simulation_reshape/Final_Simulation_GCLSTM.csv')
pedal_wiki_GSO_GCLSTM = pd.read_csv('./simulation_results/Real_simulation_reshape/Final_Simulation_GCLSTM_pedal_wiki_GSO.csv')df_GCLSTM['model']='GCLSTM'
pedal_wiki_GSO_GCLSTM['model']='GCLSTM'df_GCLSTM['dataset'].unique()array(['fivenodes', 'chickenpox', 'pedalme', 'wikimath', 'windmillsmall',
'monte'], dtype=object)- DCRNN
df_DCRNN = pd.read_csv('./simulation_results/Real_simulation_reshape/Final_Simulation_DCRNN.csv')
pedal_wiki_GSO_DCRNN = pd.read_csv('./simulation_results/Real_simulation_reshape/Final_Simulation_DCRNN_pedal_wiki_GSO.csv')df_DCRNN['model']='DCRNN'
pedal_wiki_GSO_DCRNN['model']='DCRNN'- LRGCN
df_LRGCN = pd.read_csv('./simulation_results/Real_simulation_reshape/Final_Simulation_LRGCN.csv')
pedal_wiki_GSO_LRGCN = pd.read_csv('./simulation_results/Real_simulation_reshape/Final_Simulation_LRGCN_pedal_wiki_GSO.csv')df_LRGCN['model']='LRGCN'
pedal_wiki_GSO_LRGCN['model']='LRGCN'- TGCN
df_TGCN = pd.read_csv('./simulation_results/Real_simulation_reshape/Final_Simulation_TGCN.csv')
pedal_wiki_GSO_TGCN = pd.read_csv('./simulation_results/Real_simulation_reshape/Final_Simulation_TGCN_pedal_wiki_GSO.csv')df_TGCN['model']='TGCN'
pedal_wiki_GSO_TGCN['model']='TGCN'- EvolveGCNO
df_EvolveGCNO = pd.read_csv('./simulation_results/Real_simulation_reshape/Final_Simulation_EvolveGCNO.csv')
pedal_wiki_GSO_EvolveGCNO = pd.read_csv('./simulation_results/Real_simulation_reshape/Final_Simulation_EvolveGCNO_pedal_wiki_GSO.csv')df_EvolveGCNO['model']='EvolveGCNO'
pedal_wiki_GSO_EvolveGCNO['model']='EvolveGCNO'- DYGRENCODER
df_DYGRENCODER = pd.read_csv('./simulation_results/Real_simulation_reshape/Final_Simulation_DYGRENCODER.csv')
pedal_wiki_GSO_DYGRENCODER = pd.read_csv('./simulation_results/Real_simulation_reshape/Final_Simulation_DYGRENCODER_pedal_wiki_GSO.csv')df_DYGRENCODER['model']='DyGrEncoder'
pedal_wiki_GSO_DYGRENCODER['model']='DyGrEncoder'- EvolveGCNH
df_EvolveGCNH = pd.read_csv('./simulation_results/Real_simulation_reshape/Final_Simulation_EvolveGCNH.csv')
pedal_wiki_GSO_EvolveGCNH = pd.read_csv('./simulation_results/Real_simulation_reshape/Final_Simulation_EvolveGCNH_pedal_wiki_GSO.csv')df_EvolveGCNH['model']='EvolveGCNH'
pedal_wiki_GSO_EvolveGCNH['model']='EvolveGCNH'- GNAR
df_GNAR = pd.read_csv('./simulation_results/Real_simulation_reshape/Final_Simulation_GNAR.csv')
wiki_GSO_GNAR = pd.read_csv('./simulation_results/Real_simulation_reshape/Final_Simulation_GNAR_wiki_GSO.csv')df_GNAR['model']='GNAR'
wiki_GSO_GNAR['model']='GNAR'- all
df = pd.concat([df_GConvGRU, df_GConvLSTM, df_GCLSTM, df_DCRNN, df_LRGCN, df_TGCN, df_EvolveGCNO, df_DYGRENCODER, df_EvolveGCNH,df_GNAR]).reset_index().iloc[:,1:]df| dataset | method | mrate | mtype | lags | nof_filters | inter_method | epoch | mse | calculation_time | model | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | fivenodes | STGCN | 0.0 | NaN | 2 | 12.0 | NaN | 50.0 | 0.729374 | 80.985221 | GConvGRU |
| 1 | fivenodes | STGCN | 0.0 | NaN | 2 | 12.0 | NaN | 50.0 | 0.729082 | 80.891788 | GConvGRU |
| 2 | fivenodes | STGCN | 0.7 | rand | 2 | 12.0 | linear | 50.0 | 1.892262 | 81.976547 | GConvGRU |
| 3 | fivenodes | STGCN | 0.7 | rand | 2 | 12.0 | nearest | 50.0 | 2.211288 | 87.803869 | GConvGRU |
| 4 | fivenodes | STGCN | 0.8 | rand | 2 | 12.0 | linear | 50.0 | 2.072818 | 103.648742 | GConvGRU |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 26786 | pedalme | GNAR | 0.8 | rand | 4 | NaN | nearest | NaN | 1.302679 | 0.023105 | GNAR |
| 26787 | pedalme | GNAR | 0.5 | rand | 4 | NaN | linear | NaN | 1.302679 | 0.022839 | GNAR |
| 26788 | pedalme | GNAR | 0.5 | rand | 4 | NaN | nearest | NaN | 1.302679 | 0.023041 | GNAR |
| 26789 | pedalme | GNAR | 0.8 | rand | 4 | NaN | linear | NaN | 1.302679 | 0.060872 | GNAR |
| 26790 | pedalme | GNAR | 0.8 | rand | 4 | NaN | nearest | NaN | 1.302679 | 0.023526 | GNAR |
26791 rows × 11 columns
df.to_csv('./df_fig.csv')df2 = pd.concat([pedal_wiki_GSO_GConvGRU,pedal_wiki_GSO_GConvLSTM,pedal_wiki_GSO_GCLSTM,pedal_wiki_GSO_DCRNN,pedal_wiki_GSO_LRGCN,pedal_wiki_GSO_TGCN,pedal_wiki_GSO_EvolveGCNO,pedal_wiki_GSO_DYGRENCODER,pedal_wiki_GSO_EvolveGCNH,wiki_GSO_GNAR])df2| dataset | method | mrate | mtype | lags | nof_filters | inter_method | epoch | mse | calculation_time | model | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | pedalme | STGCN | 0.300000 | rand | 4 | 12.0 | linear | 50.0 | 1.557424 | 6.553443 | GConvGRU |
| 1 | pedalme | STGCN | 0.300000 | rand | 4 | 12.0 | nearest | 50.0 | 1.591089 | 7.995071 | GConvGRU |
| 2 | pedalme | STGCN | 0.600000 | rand | 4 | 12.0 | linear | 50.0 | 1.737521 | 7.022613 | GConvGRU |
| 3 | pedalme | STGCN | 0.600000 | rand | 4 | 12.0 | nearest | 50.0 | 1.675156 | 6.745583 | GConvGRU |
| 4 | pedalme | IT-STGCN | 0.300000 | rand | 4 | 12.0 | linear | 50.0 | 1.123114 | 12.278102 | GConvGRU |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 418 | wikimath | STGCN | 0.511945 | block | 8 | 12.0 | linear | 50.0 | 0.830234 | 255.170347 | EvolveGCNH |
| 419 | wikimath | IT-STGCN | 0.511945 | block | 8 | 12.0 | linear | 50.0 | 0.746624 | 630.021935 | EvolveGCNH |
| 0 | wikimath | GNAR | 0.511945 | block | 8 | NaN | nearest | NaN | 1.353637 | 143.252073 | GNAR |
| 1 | wikimath | GNAR | 0.511945 | block | 8 | NaN | nearest | NaN | 1.353637 | 211.333467 | GNAR |
| 2 | wikimath | GNAR | 0.511945 | block | 8 | NaN | nearest | NaN | 1.353637 | 218.516156 | GNAR |
3783 rows × 11 columns
fivenode
Baseline
df.query("model=='GNAR' and dataset=='fivenodes'")| dataset | method | mrate | mtype | lags | nof_filters | inter_method | epoch | mse | calculation_time | model | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 26500 | fivenodes | GNAR | 0.0 | NaN | 2 | NaN | NaN | NaN | 1.40683 | 0.021981 | GNAR |
| 26501 | fivenodes | GNAR | 0.0 | NaN | 2 | NaN | NaN | NaN | 1.40683 | 0.017151 | GNAR |
| 26502 | fivenodes | GNAR | 0.7 | rand | 2 | NaN | linear | NaN | 1.40683 | 0.084960 | GNAR |
| 26503 | fivenodes | GNAR | 0.7 | rand | 2 | NaN | nearest | NaN | 1.40683 | 0.010853 | GNAR |
| 26504 | fivenodes | GNAR | 0.8 | rand | 2 | NaN | linear | NaN | 1.40683 | 0.012061 | GNAR |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 26768 | fivenodes | GNAR | 0.3 | rand | 2 | NaN | nearest | NaN | 1.40683 | 0.008497 | GNAR |
| 26769 | fivenodes | GNAR | 0.5 | rand | 2 | NaN | linear | NaN | 1.40683 | 0.010377 | GNAR |
| 26770 | fivenodes | GNAR | 0.5 | rand | 2 | NaN | nearest | NaN | 1.40683 | 0.018586 | GNAR |
| 26771 | fivenodes | GNAR | 0.6 | rand | 2 | NaN | linear | NaN | 1.40683 | 0.007493 | GNAR |
| 26772 | fivenodes | GNAR | 0.6 | rand | 2 | NaN | nearest | NaN | 1.40683 | 0.008042 | GNAR |
204 rows × 11 columns
pd.merge(df.query("dataset=='fivenodes' and mtype!='rand' and mtype!='block'").groupby(['model','nof_filters','lags','epoch'])['mse'].mean().reset_index(),
df.query("dataset=='fivenodes' and mtype!='rand' and mtype!='block'").groupby(['model','nof_filters','lags','epoch'])['mse'].std().reset_index(),
on=['model','nof_filters','lags','epoch']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3)| model | nof_filters | lags | epoch | mean | std | |
|---|---|---|---|---|---|---|
| 0 | DCRNN | 2.0 | 2 | 50.0 | 1.229 | 0.041 |
| 1 | DyGrEncoder | 12.0 | 2 | 50.0 | 1.114 | 0.037 |
| 2 | EvolveGCNH | 12.0 | 2 | 50.0 | 1.175 | 0.068 |
| 3 | EvolveGCNO | 12.0 | 2 | 50.0 | 1.168 | 0.065 |
| 4 | GCLSTM | 4.0 | 2 | 50.0 | 1.209 | 0.023 |
| 5 | GConvGRU | 12.0 | 2 | 50.0 | 0.732 | 0.005 |
| 6 | GConvLSTM | 12.0 | 2 | 50.0 | 1.131 | 0.041 |
| 7 | LRGCN | 4.0 | 2 | 50.0 | 1.212 | 0.024 |
| 8 | TGCN | 12.0 | 2 | 50.0 | 1.085 | 0.016 |
Random
https://matplotlib.org/stable/gallery/statistics/boxplot.html#sphx-glr-gallery-statistics-boxplot-py
# with plt.style.context('cyberpunk'):
# plt.rcParams['figure.figsize'] = [40,20]
fig, ax = plt.subplots(3, 3,figsize=(40,20))
df.query("dataset=='fivenodes' and mtype=='rand' and inter_method == 'linear' and nof_filters==12 and lags==2 and epoch==50 and model=='GConvGRU' and mrate in [0.3 , 0.5 , 0.6 ,0.7 , 0.8]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[0,0],grid=False,widths=0.5)
ax[0,0].set_title('GConvGRU')
df.query("dataset=='fivenodes' and mtype=='rand' and inter_method == 'linear' and nof_filters==12 and lags==2 and epoch==50 and model=='GConvLSTM' and mrate in [0.3 , 0.5 , 0.6 ,0.7 , 0.8]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[0,1],grid=False,widths=0.5)
ax[0,1].set_title('GConvLSTM')
df.query("dataset=='fivenodes' and mtype=='rand' and inter_method == 'linear' and nof_filters==4 and lags==2 and epoch==50 and model=='GCLSTM' and mrate in [0.3 , 0.5 , 0.6 ,0.7 , 0.8]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[0,2],grid=False,widths=0.5)
ax[0,2].set_title('GCLSTM')
df.query("dataset=='fivenodes' and mtype=='rand' and inter_method == 'linear' and nof_filters==4 and lags==2 and epoch==50 and model=='LRGCN' and mrate in [0.3 , 0.5 , 0.6 ,0.7 , 0.8]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[1,0],grid=False,widths=0.5)
ax[1,0].set_title('LRGCN')
df.query("dataset=='fivenodes' and mtype=='rand' and inter_method == 'linear' and nof_filters==12 and lags==2 and epoch==50 and model=='DyGrEncoder' and mrate in [0.3 , 0.5 , 0.6 ,0.7 , 0.8]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[1,1],grid=False,widths=0.5)
ax[1,1].set_title('DyGrEncoder')
df.query("dataset=='fivenodes' and mtype=='rand' and inter_method == 'linear' and lags==2 and epoch==50 and model=='EvolveGCNH' and mrate in [0.3 , 0.5 , 0.6 ,0.7 , 0.8]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[1,2],grid=False,widths=0.5)
ax[1,2].set_title('EvolveGCNH')
df.query("dataset=='fivenodes' and mtype=='rand' and inter_method == 'linear' and lags==2 and epoch==50 and model=='EvolveGCNO' and mrate in [0.3 , 0.5 , 0.6 ,0.7 , 0.8]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[2,0],grid=False,widths=0.5)
ax[2,0].set_title('EvolveGCNO')
df.query("dataset=='fivenodes' and mtype=='rand' and inter_method == 'linear' and nof_filters==12 and lags==2 and epoch==50 and model=='TGCN' and mrate in [0.3 , 0.5 , 0.6 ,0.7 , 0.8]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[2,1],grid=False,widths=0.5)
ax[2,1].set_title('TGCN')
df.query("dataset=='fivenodes' and mtype=='rand' and inter_method == 'linear' and nof_filters==2 and lags==2 and epoch==50 and model=='DCRNN' and mrate in [0.3 , 0.5 , 0.6 ,0.7 , 0.8]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[2,2],grid=False,widths=0.5)
ax[2,2].set_title('DCRNN')
for ax in ax.flat:
ax.set_yticklabels([])
ax.set_yscale('log')
ax.axvline(x=2.5, color='black', linestyle='-')
ax.axvline(x=4.5, color='black', linestyle='-')
ax.axvline(x=6.5, color='black', linestyle='-')
ax.axvline(x=8.5, color='black', linestyle='-')
ax.set_xticklabels(['IT-TGNN','TGNN','IT-TGNN','TGNN','IT-TGNN','TGNN','IT-TGNN','TGNN','IT-TGNN','TGNN'])
ax.set_xlabel('')
ax.set_ylabel('')
fig.suptitle('',fontsize=40)Text(0.5, 0.98, '')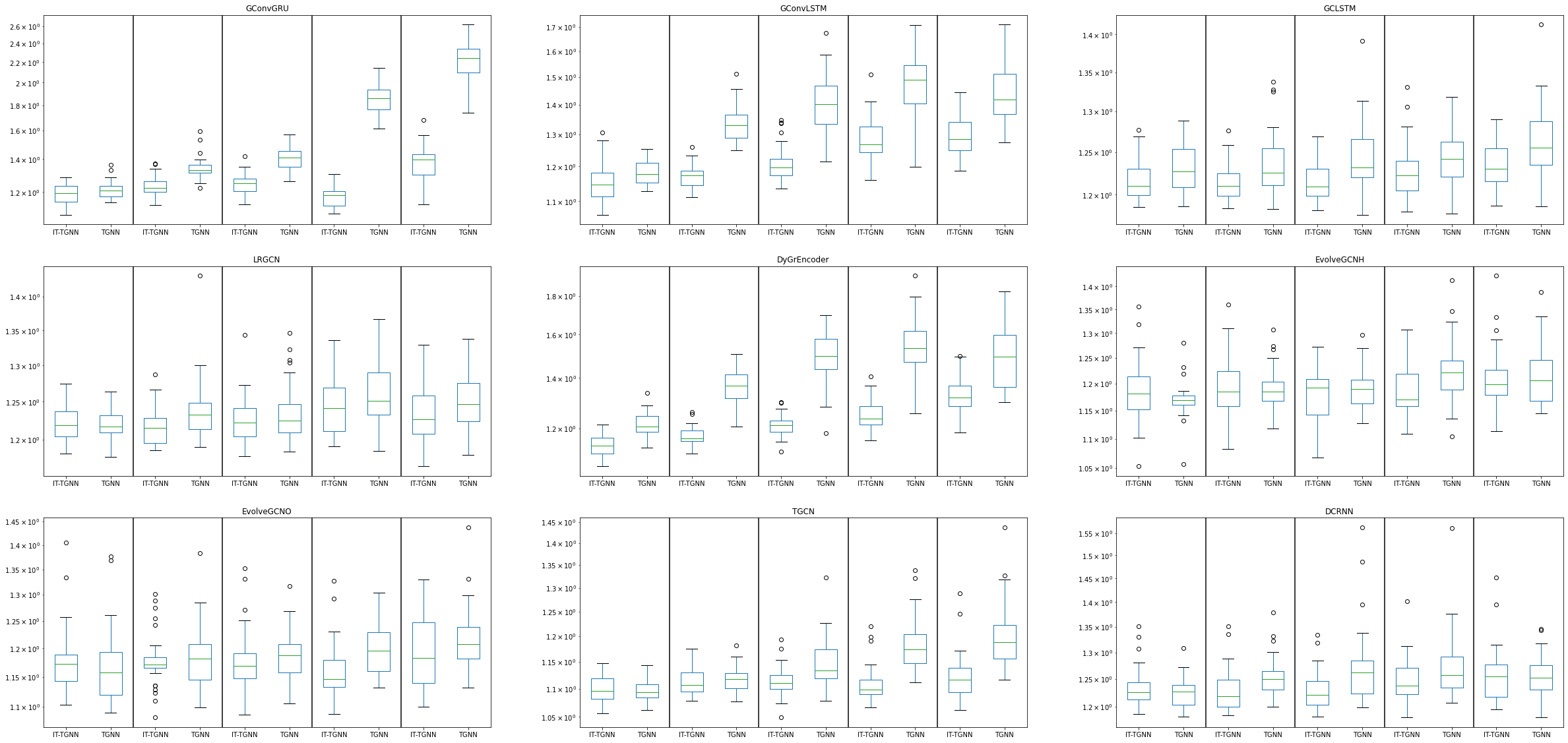
pd.merge(df.query("dataset=='fivenodes' and mtype=='rand'").groupby(['model','mrate','nof_filters','inter_method','method','lags','epoch'])['mse'].mean().reset_index(),
df.query("dataset=='fivenodes' and mtype=='rand'").groupby(['model','mrate','nof_filters','inter_method','method','lags','epoch'])['mse'].std().reset_index(),
on=['model','inter_method','method','nof_filters','mrate','lags','epoch']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3).query("mrate==0.7 and inter_method=='linear'")| model | mrate | nof_filters | inter_method | method | lags | epoch | mean | std | |
|---|---|---|---|---|---|---|---|---|---|
| 12 | DCRNN | 0.7 | 2.0 | linear | IT-STGCN | 2 | 50.0 | 1.247 | 0.044 |
| 13 | DCRNN | 0.7 | 2.0 | linear | STGCN | 2 | 50.0 | 1.271 | 0.066 |
| 32 | DyGrEncoder | 0.7 | 12.0 | linear | IT-STGCN | 2 | 50.0 | 1.252 | 0.060 |
| 33 | DyGrEncoder | 0.7 | 12.0 | linear | STGCN | 2 | 50.0 | 1.548 | 0.158 |
| 52 | EvolveGCNH | 0.7 | 12.0 | linear | IT-STGCN | 2 | 50.0 | 1.188 | 0.049 |
| 53 | EvolveGCNH | 0.7 | 12.0 | linear | STGCN | 2 | 50.0 | 1.228 | 0.064 |
| 72 | EvolveGCNO | 0.7 | 12.0 | linear | IT-STGCN | 2 | 50.0 | 1.162 | 0.052 |
| 73 | EvolveGCNO | 0.7 | 12.0 | linear | STGCN | 2 | 50.0 | 1.198 | 0.045 |
| 92 | GCLSTM | 0.7 | 4.0 | linear | IT-STGCN | 2 | 50.0 | 1.228 | 0.034 |
| 93 | GCLSTM | 0.7 | 4.0 | linear | STGCN | 2 | 50.0 | 1.245 | 0.033 |
| 112 | GConvGRU | 0.7 | 12.0 | linear | IT-STGCN | 2 | 50.0 | 1.180 | 0.060 |
| 113 | GConvGRU | 0.7 | 12.0 | linear | STGCN | 2 | 50.0 | 1.858 | 0.139 |
| 132 | GConvLSTM | 0.7 | 12.0 | linear | IT-STGCN | 2 | 50.0 | 1.287 | 0.075 |
| 133 | GConvLSTM | 0.7 | 12.0 | linear | STGCN | 2 | 50.0 | 1.472 | 0.125 |
| 152 | LRGCN | 0.7 | 4.0 | linear | IT-STGCN | 2 | 50.0 | 1.244 | 0.041 |
| 153 | LRGCN | 0.7 | 4.0 | linear | STGCN | 2 | 50.0 | 1.261 | 0.047 |
| 172 | TGCN | 0.7 | 12.0 | linear | IT-STGCN | 2 | 50.0 | 1.110 | 0.037 |
| 173 | TGCN | 0.7 | 12.0 | linear | STGCN | 2 | 50.0 | 1.184 | 0.057 |
pd.merge(df.query("dataset=='fivenodes' and mtype=='rand'").groupby(['model','mrate','nof_filters','inter_method','method','lags','epoch'])['mse'].mean().reset_index(),
df.query("dataset=='fivenodes' and mtype=='rand'").groupby(['model','mrate','nof_filters','inter_method','method','lags','epoch'])['mse'].std().reset_index(),
on=['model','inter_method','method','nof_filters','mrate','lags','epoch']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3).query("mrate==0.8")| model | mrate | nof_filters | inter_method | method | lags | epoch | mean | std | |
|---|---|---|---|---|---|---|---|---|---|
| 16 | DCRNN | 0.8 | 2.0 | linear | IT-STGCN | 2 | 50.0 | 1.257 | 0.057 |
| 17 | DCRNN | 0.8 | 2.0 | linear | STGCN | 2 | 50.0 | 1.255 | 0.040 |
| 18 | DCRNN | 0.8 | 2.0 | nearest | IT-STGCN | 2 | 50.0 | 1.246 | 0.034 |
| 19 | DCRNN | 0.8 | 2.0 | nearest | STGCN | 2 | 50.0 | 1.253 | 0.043 |
| 36 | DyGrEncoder | 0.8 | 12.0 | linear | IT-STGCN | 2 | 50.0 | 1.333 | 0.080 |
| 37 | DyGrEncoder | 0.8 | 12.0 | linear | STGCN | 2 | 50.0 | 1.496 | 0.146 |
| 38 | DyGrEncoder | 0.8 | 12.0 | nearest | IT-STGCN | 2 | 50.0 | 1.311 | 0.057 |
| 39 | DyGrEncoder | 0.8 | 12.0 | nearest | STGCN | 2 | 50.0 | 1.519 | 0.129 |
| 56 | EvolveGCNH | 0.8 | 12.0 | linear | IT-STGCN | 2 | 50.0 | 1.212 | 0.065 |
| 57 | EvolveGCNH | 0.8 | 12.0 | linear | STGCN | 2 | 50.0 | 1.217 | 0.061 |
| 58 | EvolveGCNH | 0.8 | 12.0 | nearest | IT-STGCN | 2 | 50.0 | 1.207 | 0.057 |
| 59 | EvolveGCNH | 0.8 | 12.0 | nearest | STGCN | 2 | 50.0 | 1.217 | 0.059 |
| 76 | EvolveGCNO | 0.8 | 12.0 | linear | IT-STGCN | 2 | 50.0 | 1.194 | 0.065 |
| 77 | EvolveGCNO | 0.8 | 12.0 | linear | STGCN | 2 | 50.0 | 1.220 | 0.063 |
| 78 | EvolveGCNO | 0.8 | 12.0 | nearest | IT-STGCN | 2 | 50.0 | 1.224 | 0.079 |
| 79 | EvolveGCNO | 0.8 | 12.0 | nearest | STGCN | 2 | 50.0 | 1.212 | 0.053 |
| 96 | GCLSTM | 0.8 | 4.0 | linear | IT-STGCN | 2 | 50.0 | 1.235 | 0.027 |
| 97 | GCLSTM | 0.8 | 4.0 | linear | STGCN | 2 | 50.0 | 1.263 | 0.050 |
| 98 | GCLSTM | 0.8 | 4.0 | nearest | IT-STGCN | 2 | 50.0 | 1.237 | 0.029 |
| 99 | GCLSTM | 0.8 | 4.0 | nearest | STGCN | 2 | 50.0 | 1.258 | 0.046 |
| 116 | GConvGRU | 0.8 | 12.0 | linear | IT-STGCN | 2 | 50.0 | 1.383 | 0.108 |
| 117 | GConvGRU | 0.8 | 12.0 | linear | STGCN | 2 | 50.0 | 2.224 | 0.192 |
| 118 | GConvGRU | 0.8 | 12.0 | nearest | IT-STGCN | 2 | 50.0 | 1.360 | 0.084 |
| 119 | GConvGRU | 0.8 | 12.0 | nearest | STGCN | 2 | 50.0 | 2.641 | 0.117 |
| 136 | GConvLSTM | 0.8 | 12.0 | linear | IT-STGCN | 2 | 50.0 | 1.298 | 0.060 |
| 137 | GConvLSTM | 0.8 | 12.0 | linear | STGCN | 2 | 50.0 | 1.442 | 0.111 |
| 138 | GConvLSTM | 0.8 | 12.0 | nearest | IT-STGCN | 2 | 50.0 | 1.312 | 0.065 |
| 139 | GConvLSTM | 0.8 | 12.0 | nearest | STGCN | 2 | 50.0 | 1.436 | 0.098 |
| 156 | LRGCN | 0.8 | 4.0 | linear | IT-STGCN | 2 | 50.0 | 1.233 | 0.041 |
| 157 | LRGCN | 0.8 | 4.0 | linear | STGCN | 2 | 50.0 | 1.250 | 0.038 |
| 158 | LRGCN | 0.8 | 4.0 | nearest | IT-STGCN | 2 | 50.0 | 1.230 | 0.036 |
| 159 | LRGCN | 0.8 | 4.0 | nearest | STGCN | 2 | 50.0 | 1.250 | 0.046 |
| 176 | TGCN | 0.8 | 12.0 | linear | IT-STGCN | 2 | 50.0 | 1.125 | 0.047 |
| 177 | TGCN | 0.8 | 12.0 | linear | STGCN | 2 | 50.0 | 1.199 | 0.070 |
| 178 | TGCN | 0.8 | 12.0 | nearest | IT-STGCN | 2 | 50.0 | 1.139 | 0.054 |
| 179 | TGCN | 0.8 | 12.0 | nearest | STGCN | 2 | 50.0 | 1.140 | 0.044 |
Block
pd.merge(df.query("dataset=='fivenodes' and mtype=='block'").groupby(['model','mrate','nof_filters','inter_method','method','epoch'])['mse'].mean().reset_index(),
df.query("dataset=='fivenodes' and mtype=='block'").groupby(['model','mrate','nof_filters','inter_method','method','epoch'])['mse'].std().reset_index(),
on=['model','inter_method','method','nof_filters','mrate','epoch']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3)| model | mrate | nof_filters | inter_method | method | epoch | mean | std | |
|---|---|---|---|---|---|---|---|---|
| 0 | DCRNN | 0.125 | 2.0 | linear | IT-STGCN | 50.0 | 1.232 | 0.033 |
| 1 | DCRNN | 0.125 | 2.0 | linear | STGCN | 50.0 | 1.260 | 0.051 |
| 2 | DCRNN | 0.125 | 2.0 | nearest | IT-STGCN | 50.0 | 1.222 | 0.025 |
| 3 | DCRNN | 0.125 | 2.0 | nearest | STGCN | 50.0 | 1.248 | 0.039 |
| 4 | DyGrEncoder | 0.125 | 12.0 | linear | IT-STGCN | 50.0 | 1.124 | 0.035 |
| 5 | DyGrEncoder | 0.125 | 12.0 | linear | STGCN | 50.0 | 1.173 | 0.037 |
| 6 | DyGrEncoder | 0.125 | 12.0 | nearest | IT-STGCN | 50.0 | 1.128 | 0.031 |
| 7 | DyGrEncoder | 0.125 | 12.0 | nearest | STGCN | 50.0 | 1.135 | 0.033 |
| 8 | EvolveGCNH | 0.125 | 12.0 | linear | IT-STGCN | 50.0 | 1.181 | 0.055 |
| 9 | EvolveGCNH | 0.125 | 12.0 | linear | STGCN | 50.0 | 1.197 | 0.076 |
| 10 | EvolveGCNH | 0.125 | 12.0 | nearest | IT-STGCN | 50.0 | 1.159 | 0.053 |
| 11 | EvolveGCNH | 0.125 | 12.0 | nearest | STGCN | 50.0 | 1.181 | 0.037 |
| 12 | EvolveGCNO | 0.125 | 12.0 | linear | IT-STGCN | 50.0 | 1.162 | 0.040 |
| 13 | EvolveGCNO | 0.125 | 12.0 | linear | STGCN | 50.0 | 1.176 | 0.056 |
| 14 | EvolveGCNO | 0.125 | 12.0 | nearest | IT-STGCN | 50.0 | 1.167 | 0.061 |
| 15 | EvolveGCNO | 0.125 | 12.0 | nearest | STGCN | 50.0 | 1.195 | 0.065 |
| 16 | GCLSTM | 0.125 | 4.0 | linear | IT-STGCN | 50.0 | 1.219 | 0.025 |
| 17 | GCLSTM | 0.125 | 4.0 | linear | STGCN | 50.0 | 1.244 | 0.033 |
| 18 | GCLSTM | 0.125 | 4.0 | nearest | IT-STGCN | 50.0 | 1.215 | 0.022 |
| 19 | GCLSTM | 0.125 | 4.0 | nearest | STGCN | 50.0 | 1.248 | 0.040 |
| 20 | GConvGRU | 0.125 | 12.0 | linear | IT-STGCN | 50.0 | 1.165 | 0.043 |
| 21 | GConvGRU | 0.125 | 12.0 | linear | STGCN | 50.0 | 1.210 | 0.039 |
| 22 | GConvGRU | 0.125 | 12.0 | nearest | IT-STGCN | 50.0 | 1.156 | 0.042 |
| 23 | GConvGRU | 0.125 | 12.0 | nearest | STGCN | 50.0 | 1.220 | 0.032 |
| 24 | GConvLSTM | 0.125 | 12.0 | linear | IT-STGCN | 50.0 | 1.140 | 0.038 |
| 25 | GConvLSTM | 0.125 | 12.0 | linear | STGCN | 50.0 | 1.172 | 0.055 |
| 26 | GConvLSTM | 0.125 | 12.0 | nearest | IT-STGCN | 50.0 | 1.121 | 0.027 |
| 27 | GConvLSTM | 0.125 | 12.0 | nearest | STGCN | 50.0 | 1.140 | 0.058 |
| 28 | LRGCN | 0.125 | 4.0 | linear | IT-STGCN | 50.0 | 1.220 | 0.020 |
| 29 | LRGCN | 0.125 | 4.0 | linear | STGCN | 50.0 | 1.251 | 0.037 |
| 30 | LRGCN | 0.125 | 4.0 | nearest | IT-STGCN | 50.0 | 1.216 | 0.030 |
| 31 | LRGCN | 0.125 | 4.0 | nearest | STGCN | 50.0 | 1.239 | 0.034 |
| 32 | TGCN | 0.125 | 12.0 | linear | IT-STGCN | 50.0 | 1.090 | 0.015 |
| 33 | TGCN | 0.125 | 12.0 | linear | STGCN | 50.0 | 1.107 | 0.020 |
| 34 | TGCN | 0.125 | 12.0 | nearest | IT-STGCN | 50.0 | 1.091 | 0.015 |
| 35 | TGCN | 0.125 | 12.0 | nearest | STGCN | 50.0 | 1.091 | 0.011 |
pd.merge(df.query("dataset=='fivenodes' and mtype=='block'").groupby(['model','mrate','nof_filters','inter_method','method','epoch'])['mse'].mean().reset_index(),
df.query("dataset=='fivenodes' and mtype=='block'").groupby(['model','mrate','nof_filters','inter_method','method','epoch'])['mse'].std().reset_index(),
on=['model','inter_method','method','nof_filters','mrate','epoch']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3).query("inter_method=='linear'")| model | mrate | nof_filters | inter_method | method | epoch | mean | std | |
|---|---|---|---|---|---|---|---|---|
| 0 | DCRNN | 0.125 | 2.0 | linear | IT-STGCN | 50.0 | 1.232 | 0.033 |
| 1 | DCRNN | 0.125 | 2.0 | linear | STGCN | 50.0 | 1.260 | 0.051 |
| 4 | DyGrEncoder | 0.125 | 12.0 | linear | IT-STGCN | 50.0 | 1.124 | 0.035 |
| 5 | DyGrEncoder | 0.125 | 12.0 | linear | STGCN | 50.0 | 1.173 | 0.037 |
| 8 | EvolveGCNH | 0.125 | 12.0 | linear | IT-STGCN | 50.0 | 1.181 | 0.055 |
| 9 | EvolveGCNH | 0.125 | 12.0 | linear | STGCN | 50.0 | 1.197 | 0.076 |
| 12 | EvolveGCNO | 0.125 | 12.0 | linear | IT-STGCN | 50.0 | 1.162 | 0.040 |
| 13 | EvolveGCNO | 0.125 | 12.0 | linear | STGCN | 50.0 | 1.176 | 0.056 |
| 16 | GCLSTM | 0.125 | 4.0 | linear | IT-STGCN | 50.0 | 1.219 | 0.025 |
| 17 | GCLSTM | 0.125 | 4.0 | linear | STGCN | 50.0 | 1.244 | 0.033 |
| 20 | GConvGRU | 0.125 | 12.0 | linear | IT-STGCN | 50.0 | 1.165 | 0.043 |
| 21 | GConvGRU | 0.125 | 12.0 | linear | STGCN | 50.0 | 1.210 | 0.039 |
| 24 | GConvLSTM | 0.125 | 12.0 | linear | IT-STGCN | 50.0 | 1.140 | 0.038 |
| 25 | GConvLSTM | 0.125 | 12.0 | linear | STGCN | 50.0 | 1.172 | 0.055 |
| 28 | LRGCN | 0.125 | 4.0 | linear | IT-STGCN | 50.0 | 1.220 | 0.020 |
| 29 | LRGCN | 0.125 | 4.0 | linear | STGCN | 50.0 | 1.251 | 0.037 |
| 32 | TGCN | 0.125 | 12.0 | linear | IT-STGCN | 50.0 | 1.090 | 0.015 |
| 33 | TGCN | 0.125 | 12.0 | linear | STGCN | 50.0 | 1.107 | 0.020 |
ChickenpoxDatasetLoader(lags=4)
df.query("model=='GNAR' and dataset=='chickenpox'")| dataset | method | mrate | mtype | lags | nof_filters | inter_method | epoch | mse | calculation_time | model | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 26518 | chickenpox | GNAR | 0.00000 | NaN | 4 | NaN | NaN | NaN | 1.427494 | 0.087380 | GNAR |
| 26519 | chickenpox | GNAR | 0.30000 | rand | 4 | NaN | linear | NaN | 1.427494 | 0.086311 | GNAR |
| 26520 | chickenpox | GNAR | 0.80000 | rand | 4 | NaN | linear | NaN | 1.427494 | 0.101606 | GNAR |
| 26521 | chickenpox | GNAR | 0.00000 | NaN | 4 | NaN | NaN | NaN | 1.427494 | 0.100545 | GNAR |
| 26522 | chickenpox | GNAR | 0.30000 | rand | 4 | NaN | linear | NaN | 1.427494 | 0.084384 | GNAR |
| 26523 | chickenpox | GNAR | 0.80000 | rand | 4 | NaN | linear | NaN | 1.427494 | 0.095799 | GNAR |
| 26524 | chickenpox | GNAR | 0.00000 | NaN | 4 | NaN | NaN | NaN | 1.427494 | 0.135489 | GNAR |
| 26525 | chickenpox | GNAR | 0.30000 | rand | 4 | NaN | linear | NaN | 1.427494 | 0.097362 | GNAR |
| 26526 | chickenpox | GNAR | 0.80000 | rand | 4 | NaN | linear | NaN | 1.427494 | 0.108125 | GNAR |
| 26566 | chickenpox | GNAR | 0.28777 | block | 4 | NaN | linear | NaN | 1.427494 | 0.069579 | GNAR |
| 26567 | chickenpox | GNAR | 0.28777 | block | 4 | NaN | nearest | NaN | 1.427494 | 0.073124 | GNAR |
| 26568 | chickenpox | GNAR | 0.28777 | block | 4 | NaN | linear | NaN | 1.427494 | 0.070857 | GNAR |
| 26569 | chickenpox | GNAR | 0.28777 | block | 4 | NaN | nearest | NaN | 1.427494 | 0.066239 | GNAR |
| 26570 | chickenpox | GNAR | 0.28777 | block | 4 | NaN | linear | NaN | 1.427494 | 0.070568 | GNAR |
| 26571 | chickenpox | GNAR | 0.28777 | block | 4 | NaN | nearest | NaN | 1.427494 | 0.070212 | GNAR |
| 26773 | chickenpox | GNAR | 0.50000 | rand | 4 | NaN | linear | NaN | 1.427494 | 0.077680 | GNAR |
| 26774 | chickenpox | GNAR | 0.60000 | rand | 4 | NaN | linear | NaN | 1.427494 | 0.104648 | GNAR |
| 26775 | chickenpox | GNAR | 0.50000 | rand | 4 | NaN | linear | NaN | 1.427494 | 0.123352 | GNAR |
| 26776 | chickenpox | GNAR | 0.60000 | rand | 4 | NaN | linear | NaN | 1.427494 | 0.073757 | GNAR |
| 26777 | chickenpox | GNAR | 0.50000 | rand | 4 | NaN | linear | NaN | 1.427494 | 0.082407 | GNAR |
| 26778 | chickenpox | GNAR | 0.60000 | rand | 4 | NaN | linear | NaN | 1.427494 | 0.073220 | GNAR |
Baseline
pd.merge(df.query("dataset=='chickenpox' and mtype!='rand' and mtype!='block'").groupby(['model','nof_filters'])['mse'].mean().reset_index(),
df.query("dataset=='chickenpox' and mtype!='rand' and mtype!='block'").groupby(['model','nof_filters'])['mse'].std().reset_index(),
on=['model','nof_filters']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3)| model | nof_filters | mean | std | |
|---|---|---|---|---|
| 0 | DCRNN | 16.0 | 0.727 | 0.009 |
| 1 | DyGrEncoder | 12.0 | 0.906 | 0.051 |
| 2 | EvolveGCNH | 32.0 | 1.000 | 0.020 |
| 3 | EvolveGCNO | 32.0 | 0.986 | 0.018 |
| 4 | GCLSTM | 16.0 | 0.885 | 0.051 |
| 5 | GConvGRU | 16.0 | 0.752 | 0.013 |
| 6 | GConvLSTM | 32.0 | 0.959 | 0.088 |
| 7 | LRGCN | 8.0 | 0.868 | 0.047 |
| 8 | TGCN | 12.0 | 1.090 | 0.042 |
Random
fig, ax = plt.subplots(3, 3,figsize=(40,20))
df.query("dataset=='chickenpox' and mtype=='rand' and inter_method == 'linear' and nof_filters==16 and lags==4 and epoch==50 and model=='GConvGRU' and mrate in [ 0.3 , 0.5 , 0.6 , 0.8 ]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[0,0],grid=False,widths=0.5)
ax[0,0].set_title('GConvGRU')
df.query("dataset=='chickenpox' and mtype=='rand' and inter_method == 'linear' and nof_filters==32 and lags==4 and epoch==50 and model=='GConvLSTM' and mrate in [ 0.3 , 0.5 , 0.6 , 0.8 ]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[0,1],grid=False,widths=0.5)
ax[0,1].set_title('GConvLSTM')
df.query("dataset=='chickenpox' and mtype=='rand' and inter_method == 'linear' and nof_filters==16 and lags==4 and epoch==50 and model=='GCLSTM' and mrate in [ 0.3 , 0.5 , 0.6 , 0.8 ]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[0,2],grid=False,widths=0.5)
ax[0,2].set_title('GCLSTM')
df.query("dataset=='chickenpox' and mtype=='rand' and inter_method == 'linear' and nof_filters==8 and lags==4 and epoch==50 and model=='LRGCN' and mrate in [ 0.3 , 0.5 , 0.6 , 0.8 ]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[1,0],grid=False,widths=0.5)
ax[1,0].set_title('LRGCN')
df.query("dataset=='chickenpox' and mtype=='rand' and inter_method == 'linear' and nof_filters==12 and lags==4 and epoch==50 and model=='DyGrEncoder' and mrate in [ 0.3 , 0.5 , 0.6 , 0.8 ]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[1,1],grid=False,widths=0.5)
ax[1,1].set_title('DyGrEncoder')
df.query("dataset=='chickenpox' and mtype=='rand' and inter_method == 'linear' and lags==4 and epoch==50 and model=='EvolveGCNH' and mrate in [ 0.3 , 0.5 , 0.6 , 0.8 ]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[1,2],grid=False,widths=0.5)
ax[1,2].set_title('EvolveGCNH')
df.query("dataset=='chickenpox' and mtype=='rand' and inter_method == 'linear' and lags==4 and epoch==50 and model=='EvolveGCNO' and mrate in [ 0.3 , 0.5 , 0.6 , 0.8 ]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[2,0],grid=False,widths=0.5)
ax[2,0].set_title('EvolveGCNO')
df.query("dataset=='chickenpox' and mtype=='rand' and inter_method == 'linear' and nof_filters==12 and lags==4 and epoch==50 and model=='TGCN' and mrate in [ 0.3 , 0.5 , 0.6 , 0.8 ]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[2,1],grid=False,widths=0.5)
ax[2,1].set_title('TGCN')
df.query("dataset=='chickenpox' and mtype=='rand' and inter_method == 'linear' and nof_filters==16 and lags==4 and epoch==50 and model=='DCRNN' and mrate in [ 0.3 , 0.5 , 0.6 , 0.8 ]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[2,2],grid=False,widths=0.5)
ax[2,2].set_title('DCRNN')
for ax in ax.flat:
ax.set_yticklabels([])
ax.set_yscale('log')
ax.axvline(x=2.5, color='black', linestyle='-')
ax.axvline(x=4.5, color='black', linestyle='-')
ax.axvline(x=6.5, color='black', linestyle='-')
ax.axvline(x=8.5, color='black', linestyle='-')
ax.set_xticklabels(['IT-TGNN','TGNN','IT-TGNN','TGNN','IT-TGNN','TGNN','IT-TGNN','TGNN'])
ax.set_xlabel('')
ax.set_ylabel('')
fig.suptitle('',fontsize=40)Text(0.5, 0.98, '')
pd.merge(df.query("dataset=='chickenpox' and mtype=='rand'").groupby(['model','mrate','inter_method','nof_filters','method'])['mse'].mean().reset_index(),
df.query("dataset=='chickenpox' and mtype=='rand'").groupby(['model','mrate','inter_method','nof_filters','method'])['mse'].std().reset_index(),
on=['model','method','inter_method','mrate','nof_filters']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3).query("mrate==0.3")| model | mrate | inter_method | nof_filters | method | mean | std | |
|---|---|---|---|---|---|---|---|
| 0 | DCRNN | 0.3 | linear | 16.0 | IT-STGCN | 0.797 | 0.010 |
| 1 | DCRNN | 0.3 | linear | 16.0 | STGCN | 1.032 | 0.039 |
| 8 | DyGrEncoder | 0.3 | linear | 12.0 | IT-STGCN | 0.868 | 0.028 |
| 9 | DyGrEncoder | 0.3 | linear | 12.0 | STGCN | 1.080 | 0.037 |
| 16 | EvolveGCNH | 0.3 | linear | 32.0 | IT-STGCN | 1.011 | 0.019 |
| 17 | EvolveGCNH | 0.3 | linear | 32.0 | STGCN | 1.058 | 0.015 |
| 24 | EvolveGCNO | 0.3 | linear | 32.0 | IT-STGCN | 0.998 | 0.019 |
| 25 | EvolveGCNO | 0.3 | linear | 32.0 | STGCN | 1.054 | 0.011 |
| 32 | GCLSTM | 0.3 | linear | 16.0 | IT-STGCN | 0.850 | 0.022 |
| 33 | GCLSTM | 0.3 | linear | 16.0 | STGCN | 1.050 | 0.036 |
| 40 | GConvGRU | 0.3 | linear | 16.0 | IT-STGCN | 0.851 | 0.031 |
| 41 | GConvGRU | 0.3 | linear | 16.0 | STGCN | 1.087 | 0.046 |
| 48 | GConvLSTM | 0.3 | linear | 32.0 | IT-STGCN | 0.872 | 0.035 |
| 49 | GConvLSTM | 0.3 | linear | 32.0 | STGCN | 1.114 | 0.057 |
| 56 | LRGCN | 0.3 | linear | 8.0 | IT-STGCN | 0.870 | 0.035 |
| 57 | LRGCN | 0.3 | linear | 8.0 | STGCN | 1.086 | 0.029 |
| 64 | TGCN | 0.3 | linear | 12.0 | IT-STGCN | 1.042 | 0.020 |
| 65 | TGCN | 0.3 | linear | 12.0 | STGCN | 1.054 | 0.015 |
pd.merge(df.query("dataset=='chickenpox' and mtype=='rand'").groupby(['model','mrate','inter_method','nof_filters','method'])['mse'].mean().reset_index(),
df.query("dataset=='chickenpox' and mtype=='rand'").groupby(['model','mrate','inter_method','nof_filters','method'])['mse'].std().reset_index(),
on=['model','method','inter_method','mrate','nof_filters']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3).query("mrate!=0.3")| model | mrate | inter_method | nof_filters | method | mean | std | |
|---|---|---|---|---|---|---|---|
| 2 | DCRNN | 0.5 | linear | 16.0 | IT-STGCN | 0.869 | 0.018 |
| 3 | DCRNN | 0.5 | linear | 16.0 | STGCN | 1.473 | 0.058 |
| 4 | DCRNN | 0.6 | linear | 16.0 | IT-STGCN | 0.973 | 0.032 |
| 5 | DCRNN | 0.6 | linear | 16.0 | STGCN | 1.848 | 0.072 |
| 6 | DCRNN | 0.8 | linear | 16.0 | IT-STGCN | 1.467 | 0.076 |
| 7 | DCRNN | 0.8 | linear | 16.0 | STGCN | 2.287 | 0.074 |
| 10 | DyGrEncoder | 0.5 | linear | 12.0 | IT-STGCN | 0.915 | 0.029 |
| 11 | DyGrEncoder | 0.5 | linear | 12.0 | STGCN | 1.540 | 0.045 |
| 12 | DyGrEncoder | 0.6 | linear | 12.0 | IT-STGCN | 1.013 | 0.035 |
| 13 | DyGrEncoder | 0.6 | linear | 12.0 | STGCN | 1.807 | 0.068 |
| 14 | DyGrEncoder | 0.8 | linear | 12.0 | IT-STGCN | 1.399 | 0.063 |
| 15 | DyGrEncoder | 0.8 | linear | 12.0 | STGCN | 2.127 | 0.240 |
| 18 | EvolveGCNH | 0.5 | linear | 16.0 | IT-STGCN | 1.026 | 0.017 |
| 19 | EvolveGCNH | 0.5 | linear | 16.0 | STGCN | 1.122 | 0.035 |
| 20 | EvolveGCNH | 0.6 | linear | 16.0 | IT-STGCN | 1.054 | 0.024 |
| 21 | EvolveGCNH | 0.6 | linear | 16.0 | STGCN | 1.162 | 0.044 |
| 22 | EvolveGCNH | 0.8 | linear | 32.0 | IT-STGCN | 1.140 | 0.042 |
| 23 | EvolveGCNH | 0.8 | linear | 32.0 | STGCN | 1.203 | 0.061 |
| 26 | EvolveGCNO | 0.5 | linear | 16.0 | IT-STGCN | 1.020 | 0.014 |
| 27 | EvolveGCNO | 0.5 | linear | 16.0 | STGCN | 1.145 | 0.027 |
| 28 | EvolveGCNO | 0.6 | linear | 16.0 | IT-STGCN | 1.050 | 0.018 |
| 29 | EvolveGCNO | 0.6 | linear | 16.0 | STGCN | 1.205 | 0.055 |
| 30 | EvolveGCNO | 0.8 | linear | 32.0 | IT-STGCN | 1.161 | 0.054 |
| 31 | EvolveGCNO | 0.8 | linear | 32.0 | STGCN | 1.234 | 0.096 |
| 34 | GCLSTM | 0.5 | linear | 16.0 | IT-STGCN | 0.899 | 0.023 |
| 35 | GCLSTM | 0.5 | linear | 16.0 | STGCN | 1.514 | 0.050 |
| 36 | GCLSTM | 0.6 | linear | 16.0 | IT-STGCN | 1.003 | 0.030 |
| 37 | GCLSTM | 0.6 | linear | 16.0 | STGCN | 1.808 | 0.069 |
| 38 | GCLSTM | 0.8 | linear | 16.0 | IT-STGCN | 1.371 | 0.072 |
| 39 | GCLSTM | 0.8 | linear | 16.0 | STGCN | 2.172 | 0.186 |
| 42 | GConvGRU | 0.5 | linear | 16.0 | IT-STGCN | 0.958 | 0.072 |
| 43 | GConvGRU | 0.5 | linear | 16.0 | STGCN | 1.530 | 0.106 |
| 44 | GConvGRU | 0.6 | linear | 16.0 | IT-STGCN | 1.120 | 0.072 |
| 45 | GConvGRU | 0.6 | linear | 16.0 | STGCN | 1.753 | 0.181 |
| 46 | GConvGRU | 0.8 | linear | 16.0 | IT-STGCN | 1.586 | 0.199 |
| 47 | GConvGRU | 0.8 | linear | 16.0 | STGCN | 2.529 | 0.292 |
| 50 | GConvLSTM | 0.5 | linear | 32.0 | IT-STGCN | 0.901 | 0.024 |
| 51 | GConvLSTM | 0.5 | linear | 32.0 | STGCN | 1.518 | 0.063 |
| 52 | GConvLSTM | 0.6 | linear | 32.0 | IT-STGCN | 1.004 | 0.038 |
| 53 | GConvLSTM | 0.6 | linear | 32.0 | STGCN | 1.787 | 0.055 |
| 54 | GConvLSTM | 0.8 | linear | 32.0 | IT-STGCN | 1.433 | 0.080 |
| 55 | GConvLSTM | 0.8 | linear | 32.0 | STGCN | 2.522 | 0.111 |
| 58 | LRGCN | 0.5 | linear | 8.0 | IT-STGCN | 0.931 | 0.034 |
| 59 | LRGCN | 0.5 | linear | 8.0 | STGCN | 1.458 | 0.068 |
| 60 | LRGCN | 0.6 | linear | 8.0 | IT-STGCN | 1.017 | 0.029 |
| 61 | LRGCN | 0.6 | linear | 8.0 | STGCN | 1.615 | 0.134 |
| 62 | LRGCN | 0.8 | linear | 8.0 | IT-STGCN | 1.334 | 0.071 |
| 63 | LRGCN | 0.8 | linear | 8.0 | STGCN | 1.632 | 0.156 |
| 66 | TGCN | 0.5 | linear | 12.0 | IT-STGCN | 1.032 | 0.012 |
| 67 | TGCN | 0.5 | linear | 12.0 | STGCN | 1.167 | 0.018 |
| 68 | TGCN | 0.6 | linear | 12.0 | IT-STGCN | 1.054 | 0.014 |
| 69 | TGCN | 0.6 | linear | 12.0 | STGCN | 1.242 | 0.021 |
| 70 | TGCN | 0.8 | linear | 12.0 | IT-STGCN | 1.183 | 0.028 |
| 71 | TGCN | 0.8 | linear | 12.0 | STGCN | 1.466 | 0.064 |
Block
pd.merge(df.query("dataset=='chickenpox' and mtype=='block'").groupby(['model','inter_method','mrate','nof_filters','method'])['mse'].mean().reset_index(),
df.query("dataset=='chickenpox' and mtype=='block'").groupby(['model','inter_method','mrate','nof_filters','method'])['mse'].std().reset_index(),
on=['model','method','inter_method','mrate','nof_filters']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3)| model | inter_method | mrate | nof_filters | method | mean | std | |
|---|---|---|---|---|---|---|---|
| 0 | DCRNN | linear | 0.288 | 16.0 | IT-STGCN | 0.740 | 0.007 |
| 1 | DCRNN | linear | 0.288 | 16.0 | STGCN | 0.812 | 0.006 |
| 2 | DCRNN | nearest | 0.288 | 16.0 | IT-STGCN | 0.738 | 0.007 |
| 3 | DCRNN | nearest | 0.288 | 16.0 | STGCN | 0.832 | 0.009 |
| 4 | DyGrEncoder | linear | 0.288 | 12.0 | IT-STGCN | 0.899 | 0.035 |
| 5 | DyGrEncoder | linear | 0.288 | 12.0 | STGCN | 0.912 | 0.043 |
| 6 | DyGrEncoder | nearest | 0.288 | 12.0 | IT-STGCN | 0.909 | 0.043 |
| 7 | DyGrEncoder | nearest | 0.288 | 12.0 | STGCN | 0.930 | 0.035 |
| 8 | EvolveGCNH | linear | 0.288 | 32.0 | IT-STGCN | 1.007 | 0.021 |
| 9 | EvolveGCNH | linear | 0.288 | 32.0 | STGCN | 1.027 | 0.023 |
| 10 | EvolveGCNH | nearest | 0.288 | 32.0 | IT-STGCN | 1.011 | 0.018 |
| 11 | EvolveGCNH | nearest | 0.288 | 32.0 | STGCN | 1.030 | 0.017 |
| 12 | EvolveGCNO | linear | 0.288 | 32.0 | IT-STGCN | 1.002 | 0.015 |
| 13 | EvolveGCNO | linear | 0.288 | 32.0 | STGCN | 1.028 | 0.016 |
| 14 | EvolveGCNO | nearest | 0.288 | 32.0 | IT-STGCN | 0.999 | 0.022 |
| 15 | EvolveGCNO | nearest | 0.288 | 32.0 | STGCN | 1.026 | 0.015 |
| 16 | GCLSTM | linear | 0.288 | 16.0 | IT-STGCN | 0.883 | 0.045 |
| 17 | GCLSTM | linear | 0.288 | 16.0 | STGCN | 0.890 | 0.033 |
| 18 | GCLSTM | nearest | 0.288 | 16.0 | IT-STGCN | 0.901 | 0.054 |
| 19 | GCLSTM | nearest | 0.288 | 16.0 | STGCN | 0.885 | 0.042 |
| 20 | GConvGRU | linear | 0.288 | 16.0 | IT-STGCN | 0.807 | 0.016 |
| 21 | GConvGRU | linear | 0.288 | 16.0 | STGCN | 0.828 | 0.022 |
| 22 | GConvGRU | nearest | 0.288 | 16.0 | IT-STGCN | 0.824 | 0.023 |
| 23 | GConvGRU | nearest | 0.288 | 16.0 | STGCN | 0.828 | 0.022 |
| 24 | GConvLSTM | linear | 0.288 | 32.0 | IT-STGCN | 0.911 | 0.069 |
| 25 | GConvLSTM | linear | 0.288 | 32.0 | STGCN | 0.900 | 0.049 |
| 26 | GConvLSTM | nearest | 0.288 | 32.0 | IT-STGCN | 0.885 | 0.040 |
| 27 | GConvLSTM | nearest | 0.288 | 32.0 | STGCN | 0.896 | 0.054 |
| 28 | LRGCN | linear | 0.288 | 8.0 | IT-STGCN | 0.888 | 0.035 |
| 29 | LRGCN | linear | 0.288 | 8.0 | STGCN | 0.911 | 0.047 |
| 30 | LRGCN | nearest | 0.288 | 8.0 | IT-STGCN | 0.888 | 0.041 |
| 31 | LRGCN | nearest | 0.288 | 8.0 | STGCN | 0.902 | 0.039 |
| 32 | TGCN | linear | 0.288 | 12.0 | IT-STGCN | 1.065 | 0.031 |
| 33 | TGCN | linear | 0.288 | 12.0 | STGCN | 1.082 | 0.028 |
| 34 | TGCN | nearest | 0.288 | 12.0 | IT-STGCN | 1.070 | 0.028 |
| 35 | TGCN | nearest | 0.288 | 12.0 | STGCN | 1.079 | 0.027 |
pd.merge(df.query("dataset=='chickenpox' and mtype=='block'").groupby(['model','inter_method','mrate','nof_filters','method'])['mse'].mean().reset_index(),
df.query("dataset=='chickenpox' and mtype=='block'").groupby(['model','inter_method','mrate','nof_filters','method'])['mse'].std().reset_index(),
on=['model','method','inter_method','mrate','nof_filters']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3).query("inter_method=='linear'")| model | inter_method | mrate | nof_filters | method | mean | std | |
|---|---|---|---|---|---|---|---|
| 0 | DCRNN | linear | 0.288 | 16.0 | IT-STGCN | 0.740 | 0.007 |
| 1 | DCRNN | linear | 0.288 | 16.0 | STGCN | 0.812 | 0.006 |
| 4 | DyGrEncoder | linear | 0.288 | 12.0 | IT-STGCN | 0.899 | 0.035 |
| 5 | DyGrEncoder | linear | 0.288 | 12.0 | STGCN | 0.912 | 0.043 |
| 8 | EvolveGCNH | linear | 0.288 | 32.0 | IT-STGCN | 1.007 | 0.021 |
| 9 | EvolveGCNH | linear | 0.288 | 32.0 | STGCN | 1.027 | 0.023 |
| 12 | EvolveGCNO | linear | 0.288 | 32.0 | IT-STGCN | 1.002 | 0.015 |
| 13 | EvolveGCNO | linear | 0.288 | 32.0 | STGCN | 1.028 | 0.016 |
| 16 | GCLSTM | linear | 0.288 | 16.0 | IT-STGCN | 0.883 | 0.045 |
| 17 | GCLSTM | linear | 0.288 | 16.0 | STGCN | 0.890 | 0.033 |
| 20 | GConvGRU | linear | 0.288 | 16.0 | IT-STGCN | 0.807 | 0.016 |
| 21 | GConvGRU | linear | 0.288 | 16.0 | STGCN | 0.828 | 0.022 |
| 24 | GConvLSTM | linear | 0.288 | 32.0 | IT-STGCN | 0.911 | 0.069 |
| 25 | GConvLSTM | linear | 0.288 | 32.0 | STGCN | 0.900 | 0.049 |
| 28 | LRGCN | linear | 0.288 | 8.0 | IT-STGCN | 0.888 | 0.035 |
| 29 | LRGCN | linear | 0.288 | 8.0 | STGCN | 0.911 | 0.047 |
| 32 | TGCN | linear | 0.288 | 12.0 | IT-STGCN | 1.065 | 0.031 |
| 33 | TGCN | linear | 0.288 | 12.0 | STGCN | 1.082 | 0.028 |
PedalMeDatasetLoader (lags=4)
df.query("model=='GNAR' and dataset=='pedalme'")| dataset | method | mrate | mtype | lags | nof_filters | inter_method | epoch | mse | calculation_time | model | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 26527 | pedalme | GNAR | 0.000000 | NaN | 4 | NaN | NaN | NaN | 1.302679 | 0.031931 | GNAR |
| 26528 | pedalme | GNAR | 0.000000 | NaN | 4 | NaN | NaN | NaN | 1.302679 | 0.027079 | GNAR |
| 26529 | pedalme | GNAR | 0.300000 | rand | 4 | NaN | linear | NaN | 1.302679 | 0.036771 | GNAR |
| 26530 | pedalme | GNAR | 0.300000 | rand | 4 | NaN | nearest | NaN | 1.302679 | 0.022349 | GNAR |
| 26531 | pedalme | GNAR | 0.600000 | rand | 4 | NaN | linear | NaN | 1.302679 | 0.022278 | GNAR |
| 26532 | pedalme | GNAR | 0.600000 | rand | 4 | NaN | nearest | NaN | 1.302679 | 0.029977 | GNAR |
| 26533 | pedalme | GNAR | 0.000000 | NaN | 4 | NaN | NaN | NaN | 1.302679 | 0.022007 | GNAR |
| 26534 | pedalme | GNAR | 0.000000 | NaN | 4 | NaN | NaN | NaN | 1.302679 | 0.026209 | GNAR |
| 26535 | pedalme | GNAR | 0.300000 | rand | 4 | NaN | linear | NaN | 1.302679 | 0.024794 | GNAR |
| 26536 | pedalme | GNAR | 0.300000 | rand | 4 | NaN | nearest | NaN | 1.302679 | 0.020568 | GNAR |
| 26537 | pedalme | GNAR | 0.600000 | rand | 4 | NaN | linear | NaN | 1.302679 | 0.022407 | GNAR |
| 26538 | pedalme | GNAR | 0.600000 | rand | 4 | NaN | nearest | NaN | 1.302679 | 0.021174 | GNAR |
| 26539 | pedalme | GNAR | 0.000000 | NaN | 4 | NaN | NaN | NaN | 1.302679 | 0.035516 | GNAR |
| 26540 | pedalme | GNAR | 0.000000 | NaN | 4 | NaN | NaN | NaN | 1.302679 | 0.020410 | GNAR |
| 26541 | pedalme | GNAR | 0.300000 | rand | 4 | NaN | linear | NaN | 1.302679 | 0.035666 | GNAR |
| 26542 | pedalme | GNAR | 0.300000 | rand | 4 | NaN | nearest | NaN | 1.302679 | 0.021164 | GNAR |
| 26543 | pedalme | GNAR | 0.600000 | rand | 4 | NaN | linear | NaN | 1.302679 | 0.020147 | GNAR |
| 26544 | pedalme | GNAR | 0.600000 | rand | 4 | NaN | nearest | NaN | 1.302679 | 0.021981 | GNAR |
| 26572 | pedalme | GNAR | 0.285714 | block | 4 | NaN | linear | NaN | 1.302679 | 0.017945 | GNAR |
| 26573 | pedalme | GNAR | 0.285714 | block | 4 | NaN | nearest | NaN | 1.302679 | 0.021973 | GNAR |
| 26574 | pedalme | GNAR | 0.285714 | block | 4 | NaN | linear | NaN | 1.302679 | 0.023076 | GNAR |
| 26575 | pedalme | GNAR | 0.285714 | block | 4 | NaN | nearest | NaN | 1.302679 | 0.022257 | GNAR |
| 26576 | pedalme | GNAR | 0.285714 | block | 4 | NaN | linear | NaN | 1.302679 | 0.023361 | GNAR |
| 26577 | pedalme | GNAR | 0.285714 | block | 4 | NaN | nearest | NaN | 1.302679 | 0.022614 | GNAR |
| 26779 | pedalme | GNAR | 0.500000 | rand | 4 | NaN | linear | NaN | 1.302679 | 0.038041 | GNAR |
| 26780 | pedalme | GNAR | 0.500000 | rand | 4 | NaN | nearest | NaN | 1.302679 | 0.022290 | GNAR |
| 26781 | pedalme | GNAR | 0.800000 | rand | 4 | NaN | linear | NaN | 1.302679 | 0.019467 | GNAR |
| 26782 | pedalme | GNAR | 0.800000 | rand | 4 | NaN | nearest | NaN | 1.302679 | 0.018487 | GNAR |
| 26783 | pedalme | GNAR | 0.500000 | rand | 4 | NaN | linear | NaN | 1.302679 | 0.022977 | GNAR |
| 26784 | pedalme | GNAR | 0.500000 | rand | 4 | NaN | nearest | NaN | 1.302679 | 0.039169 | GNAR |
| 26785 | pedalme | GNAR | 0.800000 | rand | 4 | NaN | linear | NaN | 1.302679 | 0.023175 | GNAR |
| 26786 | pedalme | GNAR | 0.800000 | rand | 4 | NaN | nearest | NaN | 1.302679 | 0.023105 | GNAR |
| 26787 | pedalme | GNAR | 0.500000 | rand | 4 | NaN | linear | NaN | 1.302679 | 0.022839 | GNAR |
| 26788 | pedalme | GNAR | 0.500000 | rand | 4 | NaN | nearest | NaN | 1.302679 | 0.023041 | GNAR |
| 26789 | pedalme | GNAR | 0.800000 | rand | 4 | NaN | linear | NaN | 1.302679 | 0.060872 | GNAR |
| 26790 | pedalme | GNAR | 0.800000 | rand | 4 | NaN | nearest | NaN | 1.302679 | 0.023526 | GNAR |
Baseline
pd.merge(df.query("dataset=='pedalme' and mtype!='rand' and mtype!='block'").groupby(['model','lags','nof_filters'])['mse'].mean().reset_index(),
df.query("dataset=='pedalme' and mtype!='rand' and mtype!='block'").groupby(['model','lags','nof_filters'])['mse'].std().reset_index(),
on=['model','lags','nof_filters']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3).query("lags==4")| model | lags | nof_filters | mean | std | |
|---|---|---|---|---|---|
| 0 | DCRNN | 4 | 8.0 | 1.131 | 0.015 |
| 1 | DyGrEncoder | 4 | 12.0 | 1.190 | 0.047 |
| 2 | EvolveGCNH | 4 | 2.0 | 1.213 | 0.057 |
| 3 | EvolveGCNO | 4 | 2.0 | 1.223 | 0.051 |
| 4 | GCLSTM | 4 | 4.0 | 1.181 | 0.040 |
| 5 | GConvGRU | 4 | 12.0 | 1.233 | 0.107 |
| 6 | GConvLSTM | 4 | 2.0 | 1.214 | 0.055 |
| 7 | LRGCN | 4 | 8.0 | 1.191 | 0.054 |
| 8 | TGCN | 4 | 12.0 | 1.307 | 0.075 |
Random
fig, ax = plt.subplots(3, 3,figsize=(40,20))
df.query("dataset=='pedalme' and mtype=='rand' and inter_method == 'linear' and nof_filters==12 and lags==4 and epoch==50 and model=='GConvGRU' and mrate in [0.3,0.5,0.6,0.8]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[0,0],grid=False,widths=0.5)
ax[0,0].set_title('GConvGRU')
df.query("dataset=='pedalme' and mtype=='rand' and inter_method == 'linear' and nof_filters==2 and lags==4 and epoch==50 and model=='GConvLSTM' and mrate in [0.3,0.5,0.6,0.8]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[0,1],grid=False,widths=0.5)
ax[0,1].set_title('GConvLSTM')
df.query("dataset=='pedalme' and mtype=='rand' and inter_method == 'linear' and nof_filters==4 and lags==4 and epoch==50 and model=='GCLSTM' and mrate in [0.3,0.5,0.6,0.8]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[0,2],grid=False,widths=0.5)
ax[0,2].set_title('GCLSTM')
df.query("dataset=='pedalme' and mtype=='rand' and inter_method == 'linear' and nof_filters==8 and lags==4 and epoch==50 and model=='LRGCN' and mrate in [0.3,0.5,0.6,0.8]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[1,0],grid=False,widths=0.5)
ax[1,0].set_title('LRGCN')
df.query("dataset=='pedalme' and mtype=='rand' and inter_method == 'linear' and nof_filters==12 and lags==4 and epoch==50 and model=='DyGrEncoder' and mrate in [0.3,0.5,0.6,0.8]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[1,1],grid=False,widths=0.5)
ax[1,1].set_title('DyGrEncoder')
df.query("dataset=='pedalme' and mtype=='rand' and inter_method == 'linear' and lags==4 and epoch==50 and model=='EvolveGCNH' and mrate in [0.3,0.5,0.6,0.8]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[1,2],grid=False,widths=0.5)
ax[1,2].set_title('EvolveGCNH')
df.query("dataset=='pedalme' and mtype=='rand' and inter_method == 'linear' and lags==4 and epoch==50 and model=='EvolveGCNO' and mrate in [0.3,0.5,0.6,0.8]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[2,0],grid=False,widths=0.5)
ax[2,0].set_title('EvolveGCNO')
df.query("dataset=='pedalme' and mtype=='rand' and inter_method == 'linear' and nof_filters==12 and lags==4 and epoch==50 and model=='TGCN' and mrate in [0.3,0.5,0.6,0.8]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[2,1],grid=False,widths=0.5)
ax[2,1].set_title('TGCN')
df.query("dataset=='pedalme' and mtype=='rand' and inter_method == 'linear' and nof_filters==8 and lags==4 and epoch==50 and model=='DCRNN' and mrate in [0.3,0.5,0.6,0.8]").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[2,2],grid=False,widths=0.5)
ax[2,2].set_title('DCRNN')
for ax in ax.flat:
ax.set_yticklabels([])
ax.set_yscale('log')
ax.axvline(x=2.5, color='black', linestyle='-')
ax.axvline(x=4.5, color='black', linestyle='-')
ax.axvline(x=6.5, color='black', linestyle='-')
ax.axvline(x=8.5, color='black', linestyle='-')
ax.set_xticklabels(['IT-TGNN','TGNN','IT-TGNN','TGNN','IT-TGNN','TGNN','IT-TGNN','TGNN'])
ax.set_xlabel('')
ax.set_ylabel('')
fig.suptitle('',fontsize=40)Text(0.5, 0.98, '')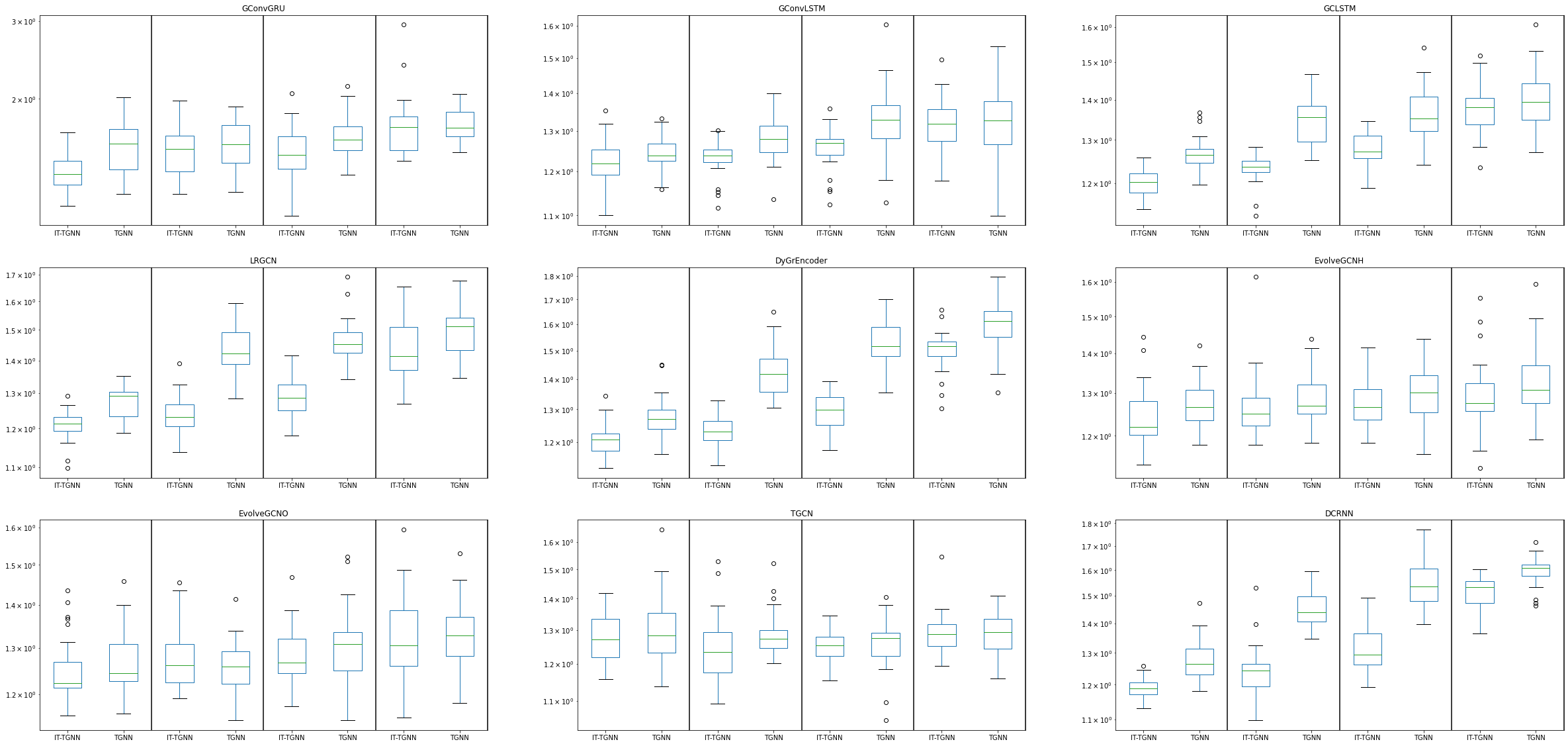
pd.merge(df.query("dataset=='pedalme' and mtype=='rand'").groupby(['model','mrate','lags','nof_filters','inter_method','method'])['mse'].mean().reset_index(),
df.query("dataset=='pedalme' and mtype=='rand'").groupby(['model','mrate','lags','nof_filters','inter_method','method'])['mse'].std().reset_index(),
on=['model','method','nof_filters','mrate','lags','inter_method']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3).query("mrate == 0.3")| model | mrate | lags | nof_filters | inter_method | method | mean | std | |
|---|---|---|---|---|---|---|---|---|
| 0 | DCRNN | 0.3 | 4 | 8.0 | linear | IT-STGCN | 1.190 | 0.029 |
| 1 | DCRNN | 0.3 | 4 | 8.0 | linear | STGCN | 1.277 | 0.064 |
| 2 | DCRNN | 0.3 | 4 | 8.0 | nearest | IT-STGCN | 1.179 | 0.035 |
| 3 | DCRNN | 0.3 | 4 | 8.0 | nearest | STGCN | 1.278 | 0.060 |
| 16 | DyGrEncoder | 0.3 | 4 | 12.0 | linear | IT-STGCN | 1.207 | 0.046 |
| 17 | DyGrEncoder | 0.3 | 4 | 12.0 | linear | STGCN | 1.279 | 0.061 |
| 18 | DyGrEncoder | 0.3 | 4 | 12.0 | nearest | IT-STGCN | 1.205 | 0.075 |
| 19 | DyGrEncoder | 0.3 | 4 | 12.0 | nearest | STGCN | 1.289 | 0.096 |
| 32 | EvolveGCNH | 0.3 | 4 | 2.0 | linear | IT-STGCN | 1.245 | 0.069 |
| 33 | EvolveGCNH | 0.3 | 4 | 2.0 | linear | STGCN | 1.273 | 0.057 |
| 34 | EvolveGCNH | 0.3 | 4 | 2.0 | nearest | IT-STGCN | 1.238 | 0.046 |
| 35 | EvolveGCNH | 0.3 | 4 | 2.0 | nearest | STGCN | 1.244 | 0.054 |
| 48 | EvolveGCNO | 0.3 | 4 | 2.0 | linear | IT-STGCN | 1.251 | 0.072 |
| 49 | EvolveGCNO | 0.3 | 4 | 2.0 | linear | STGCN | 1.267 | 0.072 |
| 50 | EvolveGCNO | 0.3 | 4 | 2.0 | nearest | IT-STGCN | 1.251 | 0.057 |
| 51 | EvolveGCNO | 0.3 | 4 | 2.0 | nearest | STGCN | 1.265 | 0.056 |
| 64 | GCLSTM | 0.3 | 4 | 4.0 | linear | IT-STGCN | 1.202 | 0.029 |
| 65 | GCLSTM | 0.3 | 4 | 4.0 | linear | STGCN | 1.267 | 0.041 |
| 66 | GCLSTM | 0.3 | 4 | 4.0 | nearest | IT-STGCN | 1.211 | 0.039 |
| 67 | GCLSTM | 0.3 | 4 | 4.0 | nearest | STGCN | 1.256 | 0.038 |
| 80 | GConvGRU | 0.3 | 4 | 12.0 | linear | IT-STGCN | 1.354 | 0.134 |
| 81 | GConvGRU | 0.3 | 4 | 12.0 | linear | STGCN | 1.575 | 0.198 |
| 82 | GConvGRU | 0.3 | 4 | 12.0 | nearest | IT-STGCN | 1.385 | 0.173 |
| 83 | GConvGRU | 0.3 | 4 | 12.0 | nearest | STGCN | 1.527 | 0.342 |
| 96 | GConvLSTM | 0.3 | 4 | 2.0 | linear | IT-STGCN | 1.227 | 0.056 |
| 97 | GConvLSTM | 0.3 | 4 | 2.0 | linear | STGCN | 1.244 | 0.041 |
| 98 | GConvLSTM | 0.3 | 4 | 2.0 | nearest | IT-STGCN | 1.224 | 0.035 |
| 99 | GConvLSTM | 0.3 | 4 | 2.0 | nearest | STGCN | 1.266 | 0.068 |
| 112 | LRGCN | 0.3 | 4 | 8.0 | linear | IT-STGCN | 1.210 | 0.039 |
| 113 | LRGCN | 0.3 | 4 | 8.0 | linear | STGCN | 1.274 | 0.045 |
| 114 | LRGCN | 0.3 | 4 | 8.0 | nearest | IT-STGCN | 1.220 | 0.046 |
| 115 | LRGCN | 0.3 | 4 | 8.0 | nearest | STGCN | 1.287 | 0.065 |
| 128 | TGCN | 0.3 | 4 | 12.0 | linear | IT-STGCN | 1.280 | 0.070 |
| 129 | TGCN | 0.3 | 4 | 12.0 | linear | STGCN | 1.302 | 0.112 |
| 130 | TGCN | 0.3 | 4 | 12.0 | nearest | IT-STGCN | 1.248 | 0.074 |
| 131 | TGCN | 0.3 | 4 | 12.0 | nearest | STGCN | 1.291 | 0.111 |
pd.merge(df.query("dataset=='pedalme' and mtype=='rand'").groupby(['model','mrate','lags','nof_filters','inter_method','method'])['mse'].mean().reset_index(),
df.query("dataset=='pedalme' and mtype=='rand'").groupby(['model','mrate','lags','nof_filters','inter_method','method'])['mse'].std().reset_index(),
on=['model','method','nof_filters','mrate','lags','inter_method']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3).query("mrate != 0.3")| model | mrate | lags | nof_filters | inter_method | method | mean | std | |
|---|---|---|---|---|---|---|---|---|
| 4 | DCRNN | 0.5 | 4 | 8.0 | linear | IT-STGCN | 1.238 | 0.080 |
| 5 | DCRNN | 0.5 | 4 | 8.0 | linear | STGCN | 1.451 | 0.061 |
| 6 | DCRNN | 0.5 | 4 | 8.0 | nearest | IT-STGCN | 1.232 | 0.061 |
| 7 | DCRNN | 0.5 | 4 | 8.0 | nearest | STGCN | 1.447 | 0.073 |
| 8 | DCRNN | 0.6 | 4 | 8.0 | linear | IT-STGCN | 1.314 | 0.072 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 139 | TGCN | 0.6 | 4 | 12.0 | nearest | STGCN | 1.301 | 0.090 |
| 140 | TGCN | 0.8 | 4 | 12.0 | linear | IT-STGCN | 1.294 | 0.063 |
| 141 | TGCN | 0.8 | 4 | 12.0 | linear | STGCN | 1.289 | 0.065 |
| 142 | TGCN | 0.8 | 4 | 12.0 | nearest | IT-STGCN | 1.258 | 0.053 |
| 143 | TGCN | 0.8 | 4 | 12.0 | nearest | STGCN | 1.277 | 0.080 |
108 rows × 8 columns
pd.merge(df.query("dataset=='pedalme' and mtype=='rand'").groupby(['model','mrate','lags','inter_method','method'])['mse'].mean().reset_index(),
df.query("dataset=='pedalme' and mtype=='rand'").groupby(['model','mrate','lags','inter_method','method'])['mse'].std().reset_index(),
on=['model','method','mrate','lags','inter_method']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3).query("mrate != 0.3").\
query("inter_method=='nearest'")| model | mrate | lags | inter_method | method | mean | std | |
|---|---|---|---|---|---|---|---|
| 6 | DCRNN | 0.5 | 4 | nearest | IT-STGCN | 1.232 | 0.061 |
| 7 | DCRNN | 0.5 | 4 | nearest | STGCN | 1.447 | 0.073 |
| 10 | DCRNN | 0.6 | 4 | nearest | IT-STGCN | 1.303 | 0.078 |
| 11 | DCRNN | 0.6 | 4 | nearest | STGCN | 1.509 | 0.068 |
| 14 | DCRNN | 0.8 | 4 | nearest | IT-STGCN | 1.527 | 0.079 |
| 15 | DCRNN | 0.8 | 4 | nearest | STGCN | 1.616 | 0.075 |
| 22 | DyGrEncoder | 0.5 | 4 | nearest | IT-STGCN | 1.236 | 0.059 |
| 23 | DyGrEncoder | 0.5 | 4 | nearest | STGCN | 1.427 | 0.076 |
| 26 | DyGrEncoder | 0.6 | 4 | nearest | IT-STGCN | 1.285 | 0.051 |
| 27 | DyGrEncoder | 0.6 | 4 | nearest | STGCN | 1.513 | 0.083 |
| 30 | DyGrEncoder | 0.8 | 4 | nearest | IT-STGCN | 1.476 | 0.098 |
| 31 | DyGrEncoder | 0.8 | 4 | nearest | STGCN | 1.614 | 0.096 |
| 38 | EvolveGCNH | 0.5 | 4 | nearest | IT-STGCN | 1.242 | 0.058 |
| 39 | EvolveGCNH | 0.5 | 4 | nearest | STGCN | 1.285 | 0.049 |
| 42 | EvolveGCNH | 0.6 | 4 | nearest | IT-STGCN | 1.262 | 0.091 |
| 43 | EvolveGCNH | 0.6 | 4 | nearest | STGCN | 1.284 | 0.066 |
| 46 | EvolveGCNH | 0.8 | 4 | nearest | IT-STGCN | 1.299 | 0.082 |
| 47 | EvolveGCNH | 0.8 | 4 | nearest | STGCN | 1.292 | 0.074 |
| 54 | EvolveGCNO | 0.5 | 4 | nearest | IT-STGCN | 1.254 | 0.037 |
| 55 | EvolveGCNO | 0.5 | 4 | nearest | STGCN | 1.274 | 0.080 |
| 58 | EvolveGCNO | 0.6 | 4 | nearest | IT-STGCN | 1.267 | 0.067 |
| 59 | EvolveGCNO | 0.6 | 4 | nearest | STGCN | 1.292 | 0.075 |
| 62 | EvolveGCNO | 0.8 | 4 | nearest | IT-STGCN | 1.312 | 0.073 |
| 63 | EvolveGCNO | 0.8 | 4 | nearest | STGCN | 1.334 | 0.144 |
| 70 | GCLSTM | 0.5 | 4 | nearest | IT-STGCN | 1.221 | 0.042 |
| 71 | GCLSTM | 0.5 | 4 | nearest | STGCN | 1.333 | 0.066 |
| 74 | GCLSTM | 0.6 | 4 | nearest | IT-STGCN | 1.259 | 0.042 |
| 75 | GCLSTM | 0.6 | 4 | nearest | STGCN | 1.365 | 0.064 |
| 78 | GCLSTM | 0.8 | 4 | nearest | IT-STGCN | 1.352 | 0.050 |
| 79 | GCLSTM | 0.8 | 4 | nearest | STGCN | 1.396 | 0.079 |
| 86 | GConvGRU | 0.5 | 4 | nearest | IT-STGCN | 1.507 | 0.235 |
| 87 | GConvGRU | 0.5 | 4 | nearest | STGCN | 1.673 | 0.223 |
| 90 | GConvGRU | 0.6 | 4 | nearest | IT-STGCN | 1.625 | 0.324 |
| 91 | GConvGRU | 0.6 | 4 | nearest | STGCN | 1.851 | 0.254 |
| 94 | GConvGRU | 0.8 | 4 | nearest | IT-STGCN | 1.608 | 0.243 |
| 95 | GConvGRU | 0.8 | 4 | nearest | STGCN | 1.871 | 0.214 |
| 102 | GConvLSTM | 0.5 | 4 | nearest | IT-STGCN | 1.235 | 0.042 |
| 103 | GConvLSTM | 0.5 | 4 | nearest | STGCN | 1.283 | 0.071 |
| 106 | GConvLSTM | 0.6 | 4 | nearest | IT-STGCN | 1.248 | 0.045 |
| 107 | GConvLSTM | 0.6 | 4 | nearest | STGCN | 1.274 | 0.078 |
| 110 | GConvLSTM | 0.8 | 4 | nearest | IT-STGCN | 1.287 | 0.065 |
| 111 | GConvLSTM | 0.8 | 4 | nearest | STGCN | 1.364 | 0.069 |
| 115 | GNAR | 0.5 | 4 | nearest | GNAR | 1.303 | 0.000 |
| 117 | GNAR | 0.6 | 4 | nearest | GNAR | 1.303 | 0.000 |
| 119 | GNAR | 0.8 | 4 | nearest | GNAR | 1.303 | 0.000 |
| 126 | LRGCN | 0.5 | 4 | nearest | IT-STGCN | 1.240 | 0.036 |
| 127 | LRGCN | 0.5 | 4 | nearest | STGCN | 1.379 | 0.079 |
| 130 | LRGCN | 0.6 | 4 | nearest | IT-STGCN | 1.286 | 0.033 |
| 131 | LRGCN | 0.6 | 4 | nearest | STGCN | 1.462 | 0.084 |
| 134 | LRGCN | 0.8 | 4 | nearest | IT-STGCN | 1.436 | 0.091 |
| 135 | LRGCN | 0.8 | 4 | nearest | STGCN | 1.529 | 0.071 |
| 142 | TGCN | 0.5 | 4 | nearest | IT-STGCN | 1.259 | 0.066 |
| 143 | TGCN | 0.5 | 4 | nearest | STGCN | 1.287 | 0.065 |
| 146 | TGCN | 0.6 | 4 | nearest | IT-STGCN | 1.260 | 0.072 |
| 147 | TGCN | 0.6 | 4 | nearest | STGCN | 1.301 | 0.090 |
| 150 | TGCN | 0.8 | 4 | nearest | IT-STGCN | 1.258 | 0.053 |
| 151 | TGCN | 0.8 | 4 | nearest | STGCN | 1.277 | 0.080 |
Block
pd.merge(df.query("dataset=='pedalme' and mtype=='block'").groupby(['model','mrate','lags','inter_method','method'])['mse'].mean().reset_index(),
df.query("dataset=='pedalme' and mtype=='block'").groupby(['model','mrate','lags','inter_method','method'])['mse'].std().reset_index(),
on=['model','method','mrate','lags','inter_method']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3).query("lags==4")| model | mrate | lags | inter_method | method | mean | std | |
|---|---|---|---|---|---|---|---|
| 0 | DCRNN | 0.286 | 4 | linear | IT-STGCN | 1.154 | 0.014 |
| 1 | DCRNN | 0.286 | 4 | linear | STGCN | 1.248 | 0.019 |
| 2 | DCRNN | 0.286 | 4 | nearest | IT-STGCN | 1.150 | 0.014 |
| 3 | DCRNN | 0.286 | 4 | nearest | STGCN | 1.304 | 0.021 |
| 4 | DyGrEncoder | 0.286 | 4 | linear | IT-STGCN | 1.167 | 0.040 |
| 5 | DyGrEncoder | 0.286 | 4 | linear | STGCN | 1.222 | 0.054 |
| 6 | DyGrEncoder | 0.286 | 4 | nearest | IT-STGCN | 1.165 | 0.032 |
| 7 | DyGrEncoder | 0.286 | 4 | nearest | STGCN | 1.269 | 0.066 |
| 8 | EvolveGCNH | 0.286 | 4 | linear | IT-STGCN | 1.259 | 0.085 |
| 9 | EvolveGCNH | 0.286 | 4 | linear | STGCN | 1.246 | 0.073 |
| 10 | EvolveGCNH | 0.286 | 4 | nearest | IT-STGCN | 1.222 | 0.040 |
| 11 | EvolveGCNH | 0.286 | 4 | nearest | STGCN | 1.265 | 0.072 |
| 12 | EvolveGCNO | 0.286 | 4 | linear | IT-STGCN | 1.246 | 0.034 |
| 13 | EvolveGCNO | 0.286 | 4 | linear | STGCN | 1.230 | 0.056 |
| 14 | EvolveGCNO | 0.286 | 4 | nearest | IT-STGCN | 1.245 | 0.045 |
| 15 | EvolveGCNO | 0.286 | 4 | nearest | STGCN | 1.246 | 0.035 |
| 16 | GCLSTM | 0.286 | 4 | linear | IT-STGCN | 1.182 | 0.031 |
| 17 | GCLSTM | 0.286 | 4 | linear | STGCN | 1.211 | 0.023 |
| 18 | GCLSTM | 0.286 | 4 | nearest | IT-STGCN | 1.195 | 0.029 |
| 19 | GCLSTM | 0.286 | 4 | nearest | STGCN | 1.248 | 0.019 |
| 20 | GConvGRU | 0.286 | 4 | linear | IT-STGCN | 1.329 | 0.131 |
| 21 | GConvGRU | 0.286 | 4 | linear | STGCN | 1.320 | 0.111 |
| 22 | GConvGRU | 0.286 | 4 | nearest | IT-STGCN | 1.289 | 0.115 |
| 23 | GConvGRU | 0.286 | 4 | nearest | STGCN | 1.270 | 0.114 |
| 24 | GConvLSTM | 0.286 | 4 | linear | IT-STGCN | 1.241 | 0.069 |
| 25 | GConvLSTM | 0.286 | 4 | linear | STGCN | 1.223 | 0.042 |
| 26 | GConvLSTM | 0.286 | 4 | nearest | IT-STGCN | 1.222 | 0.039 |
| 27 | GConvLSTM | 0.286 | 4 | nearest | STGCN | 1.237 | 0.046 |
| 28 | GNAR | 0.286 | 4 | linear | GNAR | 1.303 | 0.000 |
| 29 | GNAR | 0.286 | 4 | nearest | GNAR | 1.303 | 0.000 |
| 30 | LRGCN | 0.286 | 4 | linear | IT-STGCN | 1.169 | 0.040 |
| 31 | LRGCN | 0.286 | 4 | linear | STGCN | 1.204 | 0.032 |
| 32 | LRGCN | 0.286 | 4 | nearest | IT-STGCN | 1.165 | 0.035 |
| 33 | LRGCN | 0.286 | 4 | nearest | STGCN | 1.263 | 0.033 |
| 34 | TGCN | 0.286 | 4 | linear | IT-STGCN | 1.278 | 0.056 |
| 35 | TGCN | 0.286 | 4 | linear | STGCN | 1.244 | 0.071 |
| 36 | TGCN | 0.286 | 4 | nearest | IT-STGCN | 1.262 | 0.066 |
| 37 | TGCN | 0.286 | 4 | nearest | STGCN | 1.232 | 0.069 |
pd.merge(df.query("dataset=='pedalme' and mtype=='block'").groupby(['model','mrate','lags','inter_method','method'])['mse'].mean().reset_index(),
df.query("dataset=='pedalme' and mtype=='block'").groupby(['model','mrate','lags','inter_method','method'])['mse'].std().reset_index(),
on=['model','method','mrate','lags','inter_method']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3).\
query("lags==4 and inter_method=='nearest'")| model | mrate | lags | inter_method | method | mean | std | |
|---|---|---|---|---|---|---|---|
| 2 | DCRNN | 0.286 | 4 | nearest | IT-STGCN | 1.150 | 0.014 |
| 3 | DCRNN | 0.286 | 4 | nearest | STGCN | 1.304 | 0.021 |
| 6 | DyGrEncoder | 0.286 | 4 | nearest | IT-STGCN | 1.165 | 0.032 |
| 7 | DyGrEncoder | 0.286 | 4 | nearest | STGCN | 1.269 | 0.066 |
| 10 | EvolveGCNH | 0.286 | 4 | nearest | IT-STGCN | 1.222 | 0.040 |
| 11 | EvolveGCNH | 0.286 | 4 | nearest | STGCN | 1.265 | 0.072 |
| 14 | EvolveGCNO | 0.286 | 4 | nearest | IT-STGCN | 1.245 | 0.045 |
| 15 | EvolveGCNO | 0.286 | 4 | nearest | STGCN | 1.246 | 0.035 |
| 18 | GCLSTM | 0.286 | 4 | nearest | IT-STGCN | 1.195 | 0.029 |
| 19 | GCLSTM | 0.286 | 4 | nearest | STGCN | 1.248 | 0.019 |
| 22 | GConvGRU | 0.286 | 4 | nearest | IT-STGCN | 1.289 | 0.115 |
| 23 | GConvGRU | 0.286 | 4 | nearest | STGCN | 1.270 | 0.114 |
| 26 | GConvLSTM | 0.286 | 4 | nearest | IT-STGCN | 1.222 | 0.039 |
| 27 | GConvLSTM | 0.286 | 4 | nearest | STGCN | 1.237 | 0.046 |
| 29 | GNAR | 0.286 | 4 | nearest | GNAR | 1.303 | 0.000 |
| 32 | LRGCN | 0.286 | 4 | nearest | IT-STGCN | 1.165 | 0.035 |
| 33 | LRGCN | 0.286 | 4 | nearest | STGCN | 1.263 | 0.033 |
| 36 | TGCN | 0.286 | 4 | nearest | IT-STGCN | 1.262 | 0.066 |
| 37 | TGCN | 0.286 | 4 | nearest | STGCN | 1.232 | 0.069 |
W_st
pd.merge(df2.query("dataset == 'pedalme' and mtype=='rand'").groupby(['model','mrate','lags','inter_method','method'])['mse'].mean().reset_index(),
df2.query("dataset == 'pedalme' and mtype=='rand'").groupby(['model','mrate','lags','inter_method','method'])['mse'].std().reset_index(),
on=['model','method','mrate','lags','inter_method']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3).query("lags==4 and mrate==0.3")| model | mrate | lags | inter_method | method | mean | std | |
|---|---|---|---|---|---|---|---|
| 0 | DCRNN | 0.3 | 4 | linear | IT-STGCN | 1.153 | 0.036 |
| 1 | DCRNN | 0.3 | 4 | linear | STGCN | 1.263 | 0.053 |
| 2 | DCRNN | 0.3 | 4 | nearest | IT-STGCN | 1.154 | 0.038 |
| 3 | DCRNN | 0.3 | 4 | nearest | STGCN | 1.269 | 0.068 |
| 8 | DyGrEncoder | 0.3 | 4 | linear | IT-STGCN | 1.222 | 0.083 |
| 9 | DyGrEncoder | 0.3 | 4 | linear | STGCN | 1.276 | 0.058 |
| 10 | DyGrEncoder | 0.3 | 4 | nearest | IT-STGCN | 1.208 | 0.091 |
| 11 | DyGrEncoder | 0.3 | 4 | nearest | STGCN | 1.281 | 0.068 |
| 16 | EvolveGCNH | 0.3 | 4 | linear | IT-STGCN | 1.218 | 0.058 |
| 17 | EvolveGCNH | 0.3 | 4 | linear | STGCN | 1.237 | 0.051 |
| 18 | EvolveGCNH | 0.3 | 4 | nearest | IT-STGCN | 1.197 | 0.068 |
| 19 | EvolveGCNH | 0.3 | 4 | nearest | STGCN | 1.237 | 0.058 |
| 24 | EvolveGCNO | 0.3 | 4 | linear | IT-STGCN | 1.223 | 0.041 |
| 25 | EvolveGCNO | 0.3 | 4 | linear | STGCN | 1.263 | 0.048 |
| 26 | EvolveGCNO | 0.3 | 4 | nearest | IT-STGCN | 1.234 | 0.046 |
| 27 | EvolveGCNO | 0.3 | 4 | nearest | STGCN | 1.252 | 0.071 |
| 32 | GCLSTM | 0.3 | 4 | linear | IT-STGCN | 1.191 | 0.041 |
| 33 | GCLSTM | 0.3 | 4 | linear | STGCN | 1.264 | 0.041 |
| 34 | GCLSTM | 0.3 | 4 | nearest | IT-STGCN | 1.193 | 0.033 |
| 35 | GCLSTM | 0.3 | 4 | nearest | STGCN | 1.250 | 0.049 |
| 40 | GConvGRU | 0.3 | 4 | linear | IT-STGCN | 1.270 | 0.163 |
| 41 | GConvGRU | 0.3 | 4 | linear | STGCN | 1.556 | 0.264 |
| 42 | GConvGRU | 0.3 | 4 | nearest | IT-STGCN | 1.324 | 0.163 |
| 43 | GConvGRU | 0.3 | 4 | nearest | STGCN | 1.520 | 0.206 |
| 48 | GConvLSTM | 0.3 | 4 | linear | IT-STGCN | 1.340 | 0.166 |
| 49 | GConvLSTM | 0.3 | 4 | linear | STGCN | 1.392 | 0.109 |
| 50 | GConvLSTM | 0.3 | 4 | nearest | IT-STGCN | 1.368 | 0.158 |
| 51 | GConvLSTM | 0.3 | 4 | nearest | STGCN | 1.338 | 0.118 |
| 56 | LRGCN | 0.3 | 4 | linear | IT-STGCN | 1.299 | 0.147 |
| 57 | LRGCN | 0.3 | 4 | linear | STGCN | 1.325 | 0.086 |
| 58 | LRGCN | 0.3 | 4 | nearest | IT-STGCN | 1.260 | 0.117 |
| 59 | LRGCN | 0.3 | 4 | nearest | STGCN | 1.269 | 0.087 |
| 64 | TGCN | 0.3 | 4 | linear | IT-STGCN | 1.320 | 0.164 |
| 65 | TGCN | 0.3 | 4 | linear | STGCN | 1.287 | 0.126 |
| 66 | TGCN | 0.3 | 4 | nearest | IT-STGCN | 1.276 | 0.105 |
| 67 | TGCN | 0.3 | 4 | nearest | STGCN | 1.313 | 0.101 |
- 테이블 정리용
pd.merge(df2.query("dataset == 'pedalme' and mtype=='rand'").groupby(['model','mrate','lags','inter_method','method'])['mse'].mean().reset_index(),
df2.query("dataset == 'pedalme' and mtype=='rand'").groupby(['model','mrate','lags','inter_method','method'])['mse'].std().reset_index(),
on=['model','method','mrate','lags','inter_method']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3).query("lags==4 and mrate == 0.6 and inter_method=='linear'")| model | mrate | lags | inter_method | method | mean | std | |
|---|---|---|---|---|---|---|---|
| 4 | DCRNN | 0.6 | 4 | linear | IT-STGCN | 1.241 | 0.079 |
| 5 | DCRNN | 0.6 | 4 | linear | STGCN | 1.506 | 0.065 |
| 12 | DyGrEncoder | 0.6 | 4 | linear | IT-STGCN | 1.287 | 0.095 |
| 13 | DyGrEncoder | 0.6 | 4 | linear | STGCN | 1.497 | 0.077 |
| 20 | EvolveGCNH | 0.6 | 4 | linear | IT-STGCN | 1.229 | 0.070 |
| 21 | EvolveGCNH | 0.6 | 4 | linear | STGCN | 1.278 | 0.066 |
| 28 | EvolveGCNO | 0.6 | 4 | linear | IT-STGCN | 1.269 | 0.092 |
| 29 | EvolveGCNO | 0.6 | 4 | linear | STGCN | 1.304 | 0.061 |
| 36 | GCLSTM | 0.6 | 4 | linear | IT-STGCN | 1.260 | 0.084 |
| 37 | GCLSTM | 0.6 | 4 | linear | STGCN | 1.340 | 0.059 |
| 44 | GConvGRU | 0.6 | 4 | linear | IT-STGCN | 1.434 | 0.222 |
| 45 | GConvGRU | 0.6 | 4 | linear | STGCN | 1.678 | 0.211 |
| 52 | GConvLSTM | 0.6 | 4 | linear | IT-STGCN | 1.312 | 0.162 |
| 53 | GConvLSTM | 0.6 | 4 | linear | STGCN | 1.498 | 0.083 |
| 60 | LRGCN | 0.6 | 4 | linear | IT-STGCN | 1.265 | 0.100 |
| 61 | LRGCN | 0.6 | 4 | linear | STGCN | 1.466 | 0.085 |
| 68 | TGCN | 0.6 | 4 | linear | IT-STGCN | 1.304 | 0.129 |
| 69 | TGCN | 0.6 | 4 | linear | STGCN | 1.299 | 0.076 |
pd.merge(df2.query("dataset == 'pedalme' and mtype=='rand'").groupby(['model','mrate','lags','inter_method','method'])['mse'].mean().reset_index(),
df2.query("dataset == 'pedalme' and mtype=='rand'").groupby(['model','mrate','lags','inter_method','method'])['mse'].std().reset_index(),
on=['model','method','mrate','lags','inter_method']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3).query("method !='STGCN' and lags==4 and mrate == 0.6 and inter_method=='linear'")| model | mrate | lags | inter_method | method | mean | std | |
|---|---|---|---|---|---|---|---|
| 4 | DCRNN | 0.6 | 4 | linear | IT-STGCN | 1.241 | 0.079 |
| 12 | DyGrEncoder | 0.6 | 4 | linear | IT-STGCN | 1.287 | 0.095 |
| 20 | EvolveGCNH | 0.6 | 4 | linear | IT-STGCN | 1.229 | 0.070 |
| 28 | EvolveGCNO | 0.6 | 4 | linear | IT-STGCN | 1.269 | 0.092 |
| 36 | GCLSTM | 0.6 | 4 | linear | IT-STGCN | 1.260 | 0.084 |
| 44 | GConvGRU | 0.6 | 4 | linear | IT-STGCN | 1.434 | 0.222 |
| 52 | GConvLSTM | 0.6 | 4 | linear | IT-STGCN | 1.312 | 0.162 |
| 60 | LRGCN | 0.6 | 4 | linear | IT-STGCN | 1.265 | 0.100 |
| 68 | TGCN | 0.6 | 4 | linear | IT-STGCN | 1.304 | 0.129 |
pd.merge(df.query("dataset == 'pedalme' and mtype=='rand'").groupby(['model','mrate','lags','inter_method','method'])['mse'].mean().reset_index(),
df.query("dataset == 'pedalme' and mtype=='rand'").groupby(['model','mrate','lags','inter_method','method'])['mse'].std().reset_index(),
on=['model','method','mrate','lags','inter_method']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3).query("method !='STGCN' and lags==4 and mrate == 0.6 and inter_method=='linear'")| model | mrate | lags | inter_method | method | mean | std | |
|---|---|---|---|---|---|---|---|
| 8 | DCRNN | 0.6 | 4 | linear | IT-STGCN | 1.314 | 0.072 |
| 24 | DyGrEncoder | 0.6 | 4 | linear | IT-STGCN | 1.294 | 0.056 |
| 40 | EvolveGCNH | 0.6 | 4 | linear | IT-STGCN | 1.279 | 0.057 |
| 56 | EvolveGCNO | 0.6 | 4 | linear | IT-STGCN | 1.280 | 0.065 |
| 72 | GCLSTM | 0.6 | 4 | linear | IT-STGCN | 1.278 | 0.040 |
| 88 | GConvGRU | 0.6 | 4 | linear | IT-STGCN | 1.516 | 0.211 |
| 104 | GConvLSTM | 0.6 | 4 | linear | IT-STGCN | 1.255 | 0.049 |
| 116 | GNAR | 0.6 | 4 | linear | GNAR | 1.303 | 0.000 |
| 128 | LRGCN | 0.6 | 4 | linear | IT-STGCN | 1.286 | 0.053 |
| 144 | TGCN | 0.6 | 4 | linear | IT-STGCN | 1.257 | 0.048 |
pd.merge(df2.query("dataset == 'pedalme' and mtype=='rand'").groupby(['model','mrate','lags','inter_method','method'])['mse'].mean().reset_index(),
df2.query("dataset == 'pedalme' and mtype=='rand'").groupby(['model','mrate','lags','inter_method','method'])['mse'].std().reset_index(),
on=['model','method','mrate','lags','inter_method']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3).query("lags==4 and mrate == 0.6")| model | mrate | lags | inter_method | method | mean | std | |
|---|---|---|---|---|---|---|---|
| 4 | DCRNN | 0.6 | 4 | linear | IT-STGCN | 1.241 | 0.079 |
| 5 | DCRNN | 0.6 | 4 | linear | STGCN | 1.506 | 0.065 |
| 6 | DCRNN | 0.6 | 4 | nearest | IT-STGCN | 1.208 | 0.079 |
| 7 | DCRNN | 0.6 | 4 | nearest | STGCN | 1.552 | 0.087 |
| 12 | DyGrEncoder | 0.6 | 4 | linear | IT-STGCN | 1.287 | 0.095 |
| 13 | DyGrEncoder | 0.6 | 4 | linear | STGCN | 1.497 | 0.077 |
| 14 | DyGrEncoder | 0.6 | 4 | nearest | IT-STGCN | 1.305 | 0.131 |
| 15 | DyGrEncoder | 0.6 | 4 | nearest | STGCN | 1.513 | 0.073 |
| 20 | EvolveGCNH | 0.6 | 4 | linear | IT-STGCN | 1.229 | 0.070 |
| 21 | EvolveGCNH | 0.6 | 4 | linear | STGCN | 1.278 | 0.066 |
| 22 | EvolveGCNH | 0.6 | 4 | nearest | IT-STGCN | 1.246 | 0.067 |
| 23 | EvolveGCNH | 0.6 | 4 | nearest | STGCN | 1.291 | 0.063 |
| 28 | EvolveGCNO | 0.6 | 4 | linear | IT-STGCN | 1.269 | 0.092 |
| 29 | EvolveGCNO | 0.6 | 4 | linear | STGCN | 1.304 | 0.061 |
| 30 | EvolveGCNO | 0.6 | 4 | nearest | IT-STGCN | 1.248 | 0.072 |
| 31 | EvolveGCNO | 0.6 | 4 | nearest | STGCN | 1.321 | 0.094 |
| 36 | GCLSTM | 0.6 | 4 | linear | IT-STGCN | 1.260 | 0.084 |
| 37 | GCLSTM | 0.6 | 4 | linear | STGCN | 1.340 | 0.059 |
| 38 | GCLSTM | 0.6 | 4 | nearest | IT-STGCN | 1.231 | 0.044 |
| 39 | GCLSTM | 0.6 | 4 | nearest | STGCN | 1.355 | 0.068 |
| 44 | GConvGRU | 0.6 | 4 | linear | IT-STGCN | 1.434 | 0.222 |
| 45 | GConvGRU | 0.6 | 4 | linear | STGCN | 1.678 | 0.211 |
| 46 | GConvGRU | 0.6 | 4 | nearest | IT-STGCN | 1.410 | 0.208 |
| 47 | GConvGRU | 0.6 | 4 | nearest | STGCN | 1.771 | 0.220 |
| 52 | GConvLSTM | 0.6 | 4 | linear | IT-STGCN | 1.312 | 0.162 |
| 53 | GConvLSTM | 0.6 | 4 | linear | STGCN | 1.498 | 0.083 |
| 54 | GConvLSTM | 0.6 | 4 | nearest | IT-STGCN | 1.313 | 0.205 |
| 55 | GConvLSTM | 0.6 | 4 | nearest | STGCN | 1.503 | 0.101 |
| 60 | LRGCN | 0.6 | 4 | linear | IT-STGCN | 1.265 | 0.100 |
| 61 | LRGCN | 0.6 | 4 | linear | STGCN | 1.466 | 0.085 |
| 62 | LRGCN | 0.6 | 4 | nearest | IT-STGCN | 1.331 | 0.120 |
| 63 | LRGCN | 0.6 | 4 | nearest | STGCN | 1.453 | 0.115 |
| 68 | TGCN | 0.6 | 4 | linear | IT-STGCN | 1.304 | 0.129 |
| 69 | TGCN | 0.6 | 4 | linear | STGCN | 1.299 | 0.076 |
| 70 | TGCN | 0.6 | 4 | nearest | IT-STGCN | 1.338 | 0.202 |
| 71 | TGCN | 0.6 | 4 | nearest | STGCN | 1.297 | 0.093 |
- conclusion table
pd.merge(df2.query("dataset == 'pedalme' and mtype=='block'").groupby(['model','mrate','lags','inter_method','method'])['mse'].mean().reset_index(),
df2.query("dataset == 'pedalme' and mtype=='block'").groupby(['model','mrate','lags','inter_method','method'])['mse'].std().reset_index(),
on=['model','method','mrate','lags','inter_method']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3).query("lags==4 and inter_method=='linear'")| model | mrate | lags | inter_method | method | mean | std | |
|---|---|---|---|---|---|---|---|
| 0 | DCRNN | 0.286 | 4 | linear | IT-STGCN | 1.145 | 0.013 |
| 1 | DCRNN | 0.286 | 4 | linear | STGCN | 1.295 | 0.019 |
| 4 | DyGrEncoder | 0.286 | 4 | linear | IT-STGCN | 1.196 | 0.055 |
| 5 | DyGrEncoder | 0.286 | 4 | linear | STGCN | 1.224 | 0.037 |
| 8 | EvolveGCNH | 0.286 | 4 | linear | IT-STGCN | 1.188 | 0.042 |
| 9 | EvolveGCNH | 0.286 | 4 | linear | STGCN | 1.230 | 0.056 |
| 12 | EvolveGCNO | 0.286 | 4 | linear | IT-STGCN | 1.204 | 0.033 |
| 13 | EvolveGCNO | 0.286 | 4 | linear | STGCN | 1.210 | 0.058 |
| 16 | GCLSTM | 0.286 | 4 | linear | IT-STGCN | 1.182 | 0.045 |
| 17 | GCLSTM | 0.286 | 4 | linear | STGCN | 1.225 | 0.030 |
| 20 | GConvGRU | 0.286 | 4 | linear | IT-STGCN | 1.391 | 0.151 |
| 21 | GConvGRU | 0.286 | 4 | linear | STGCN | 1.420 | 0.110 |
| 24 | GConvLSTM | 0.286 | 4 | linear | IT-STGCN | 1.329 | 0.120 |
| 25 | GConvLSTM | 0.286 | 4 | linear | STGCN | 1.372 | 0.199 |
| 28 | LRGCN | 0.286 | 4 | linear | IT-STGCN | 1.201 | 0.081 |
| 29 | LRGCN | 0.286 | 4 | linear | STGCN | 1.227 | 0.070 |
| 32 | TGCN | 0.286 | 4 | linear | IT-STGCN | 1.243 | 0.110 |
| 33 | TGCN | 0.286 | 4 | linear | STGCN | 1.176 | 0.068 |
pd.merge(df2.query("dataset == 'pedalme' and mtype=='block'").groupby(['model','mrate','lags','inter_method','method'])['mse'].mean().reset_index(),
df2.query("dataset == 'pedalme' and mtype=='block'").groupby(['model','mrate','lags','inter_method','method'])['mse'].std().reset_index(),
on=['model','method','mrate','lags','inter_method']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3).query("lags==4")| model | mrate | lags | inter_method | method | mean | std | |
|---|---|---|---|---|---|---|---|
| 0 | DCRNN | 0.286 | 4 | linear | IT-STGCN | 1.145 | 0.013 |
| 1 | DCRNN | 0.286 | 4 | linear | STGCN | 1.295 | 0.019 |
| 2 | DCRNN | 0.286 | 4 | nearest | IT-STGCN | 1.143 | 0.011 |
| 3 | DCRNN | 0.286 | 4 | nearest | STGCN | 1.310 | 0.019 |
| 4 | DyGrEncoder | 0.286 | 4 | linear | IT-STGCN | 1.196 | 0.055 |
| 5 | DyGrEncoder | 0.286 | 4 | linear | STGCN | 1.224 | 0.037 |
| 6 | DyGrEncoder | 0.286 | 4 | nearest | IT-STGCN | 1.204 | 0.063 |
| 7 | DyGrEncoder | 0.286 | 4 | nearest | STGCN | 1.246 | 0.043 |
| 8 | EvolveGCNH | 0.286 | 4 | linear | IT-STGCN | 1.188 | 0.042 |
| 9 | EvolveGCNH | 0.286 | 4 | linear | STGCN | 1.230 | 0.056 |
| 10 | EvolveGCNH | 0.286 | 4 | nearest | IT-STGCN | 1.195 | 0.037 |
| 11 | EvolveGCNH | 0.286 | 4 | nearest | STGCN | 1.240 | 0.062 |
| 12 | EvolveGCNO | 0.286 | 4 | linear | IT-STGCN | 1.204 | 0.033 |
| 13 | EvolveGCNO | 0.286 | 4 | linear | STGCN | 1.210 | 0.058 |
| 14 | EvolveGCNO | 0.286 | 4 | nearest | IT-STGCN | 1.211 | 0.033 |
| 15 | EvolveGCNO | 0.286 | 4 | nearest | STGCN | 1.241 | 0.095 |
| 16 | GCLSTM | 0.286 | 4 | linear | IT-STGCN | 1.182 | 0.045 |
| 17 | GCLSTM | 0.286 | 4 | linear | STGCN | 1.225 | 0.030 |
| 18 | GCLSTM | 0.286 | 4 | nearest | IT-STGCN | 1.185 | 0.035 |
| 19 | GCLSTM | 0.286 | 4 | nearest | STGCN | 1.249 | 0.027 |
| 20 | GConvGRU | 0.286 | 4 | linear | IT-STGCN | 1.391 | 0.151 |
| 21 | GConvGRU | 0.286 | 4 | linear | STGCN | 1.420 | 0.110 |
| 22 | GConvGRU | 0.286 | 4 | nearest | IT-STGCN | 1.361 | 0.114 |
| 23 | GConvGRU | 0.286 | 4 | nearest | STGCN | 1.430 | 0.145 |
| 24 | GConvLSTM | 0.286 | 4 | linear | IT-STGCN | 1.329 | 0.120 |
| 25 | GConvLSTM | 0.286 | 4 | linear | STGCN | 1.372 | 0.199 |
| 26 | GConvLSTM | 0.286 | 4 | nearest | IT-STGCN | 1.310 | 0.151 |
| 27 | GConvLSTM | 0.286 | 4 | nearest | STGCN | 1.459 | 0.153 |
| 28 | LRGCN | 0.286 | 4 | linear | IT-STGCN | 1.201 | 0.081 |
| 29 | LRGCN | 0.286 | 4 | linear | STGCN | 1.227 | 0.070 |
| 30 | LRGCN | 0.286 | 4 | nearest | IT-STGCN | 1.197 | 0.106 |
| 31 | LRGCN | 0.286 | 4 | nearest | STGCN | 1.347 | 0.117 |
| 32 | TGCN | 0.286 | 4 | linear | IT-STGCN | 1.243 | 0.110 |
| 33 | TGCN | 0.286 | 4 | linear | STGCN | 1.176 | 0.068 |
| 34 | TGCN | 0.286 | 4 | nearest | IT-STGCN | 1.237 | 0.083 |
| 35 | TGCN | 0.286 | 4 | nearest | STGCN | 1.258 | 0.064 |
WikiMathsDatasetLoader (lags=8)
df.query("model=='GNAR' and dataset=='wikimath'")| dataset | method | mrate | mtype | lags | nof_filters | inter_method | epoch | mse | calculation_time | model | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 26545 | wikimath | GNAR | 0.000000 | NaN | 8 | NaN | NaN | NaN | 1.353637 | 228.561262 | GNAR |
| 26546 | wikimath | GNAR | 0.300000 | rand | 8 | NaN | linear | NaN | 1.353637 | 214.543727 | GNAR |
| 26547 | wikimath | GNAR | 0.800000 | rand | 8 | NaN | linear | NaN | 1.353637 | 204.452467 | GNAR |
| 26548 | wikimath | GNAR | 0.000000 | NaN | 8 | NaN | NaN | NaN | 1.353637 | 129.201734 | GNAR |
| 26549 | wikimath | GNAR | 0.300000 | rand | 8 | NaN | linear | NaN | 1.353637 | 193.721538 | GNAR |
| 26550 | wikimath | GNAR | 0.800000 | rand | 8 | NaN | linear | NaN | 1.353637 | 188.391585 | GNAR |
| 26551 | wikimath | GNAR | 0.000000 | NaN | 8 | NaN | NaN | NaN | 1.353637 | 115.926373 | GNAR |
| 26552 | wikimath | GNAR | 0.300000 | rand | 8 | NaN | linear | NaN | 1.353637 | 223.202336 | GNAR |
| 26553 | wikimath | GNAR | 0.800000 | rand | 8 | NaN | linear | NaN | 1.353637 | 126.158856 | GNAR |
| 26578 | wikimath | GNAR | 0.119837 | block | 8 | NaN | linear | NaN | 1.353637 | 114.421239 | GNAR |
| 26579 | wikimath | GNAR | 0.119837 | block | 8 | NaN | linear | NaN | 1.353637 | 214.541294 | GNAR |
| 26580 | wikimath | GNAR | 0.119837 | block | 8 | NaN | linear | NaN | 1.353637 | 269.699585 | GNAR |
Baseline
pd.merge(df.query("dataset=='wikimath' and mrate==0").groupby(['model','lags','nof_filters','method'])['mse'].mean().reset_index(),
df.query("dataset=='wikimath' and mrate==0").groupby(['model','lags','nof_filters','method'])['mse'].std().reset_index(),
on=['model','lags','nof_filters','method']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3)| model | lags | nof_filters | method | mean | std | |
|---|---|---|---|---|---|---|
| 0 | DCRNN | 8 | 12.0 | IT-STGCN | 0.582 | 0.006 |
| 1 | DCRNN | 8 | 12.0 | STGCN | 0.580 | 0.006 |
| 2 | DyGrEncoder | 8 | 12.0 | IT-STGCN | 0.563 | 0.030 |
| 3 | DyGrEncoder | 8 | 12.0 | STGCN | 0.560 | 0.029 |
| 4 | EvolveGCNH | 8 | 12.0 | IT-STGCN | 0.784 | 0.027 |
| 5 | EvolveGCNH | 8 | 12.0 | STGCN | 0.771 | 0.028 |
| 6 | EvolveGCNO | 8 | 12.0 | IT-STGCN | 0.735 | 0.023 |
| 7 | EvolveGCNO | 8 | 12.0 | STGCN | 0.734 | 0.025 |
| 8 | GCLSTM | 8 | 64.0 | IT-STGCN | 0.643 | 0.024 |
| 9 | GCLSTM | 8 | 64.0 | STGCN | 0.645 | 0.018 |
| 10 | GConvGRU | 8 | 12.0 | IT-STGCN | 0.529 | 0.003 |
| 11 | GConvGRU | 8 | 12.0 | STGCN | 0.528 | 0.003 |
| 12 | GConvLSTM | 8 | 64.0 | IT-STGCN | 0.626 | 0.015 |
| 13 | GConvLSTM | 8 | 64.0 | STGCN | 0.640 | 0.031 |
| 14 | LRGCN | 8 | 32.0 | IT-STGCN | 0.610 | 0.017 |
| 15 | LRGCN | 8 | 32.0 | STGCN | 0.608 | 0.014 |
| 16 | TGCN | 8 | 12.0 | IT-STGCN | 0.730 | 0.030 |
| 17 | TGCN | 8 | 12.0 | STGCN | 0.732 | 0.035 |
Random
fig, ax = plt.subplots(3, 3,figsize=(40,20))
df.query("dataset=='wikimath' and mtype=='rand' and inter_method == 'linear' and nof_filters==12 and lags==8 and epoch==50 and model=='GConvGRU'").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[0,0],grid=False,widths=0.5)
ax[0,0].set_title('GConvGRU')
df.query("dataset=='wikimath' and mtype=='rand' and inter_method == 'linear' and nof_filters==64 and lags==8 and epoch==50 and model=='GConvLSTM'").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[0,1],grid=False,widths=0.5)
ax[0,1].set_title('GConvLSTM')
df.query("dataset=='wikimath' and mtype=='rand' and inter_method == 'linear' and nof_filters==64 and lags==8 and epoch==50 and model=='GCLSTM'").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[0,2],grid=False,widths=0.5)
ax[0,2].set_title('GCLSTM')
df.query("dataset=='wikimath' and mtype=='rand' and inter_method == 'linear' and nof_filters==32 and lags==8 and epoch==50 and model=='LRGCN'").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[1,0],grid=False,widths=0.5)
ax[1,0].set_title('LRGCN')
df.query("dataset=='wikimath' and mtype=='rand' and inter_method == 'linear' and nof_filters==12 and lags==8 and epoch==50 and model=='DyGrEncoder'").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[1,1],grid=False,widths=0.5)
ax[1,1].set_title('DyGrEncoder')
df.query("dataset=='wikimath' and mtype=='rand' and inter_method == 'linear' and lags==8 and epoch==50 and model=='EvolveGCNH'").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[1,2],grid=False,widths=0.5)
ax[1,2].set_title('EvolveGCNH')
df.query("dataset=='wikimath' and mtype=='rand' and inter_method == 'linear' and lags==8 and epoch==50 and model=='EvolveGCNO'").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[2,0],grid=False,widths=0.5)
ax[2,0].set_title('EvolveGCNO')
df.query("dataset=='wikimath' and mtype=='rand' and inter_method == 'linear' and nof_filters==12 and lags==8 and epoch==50 and model=='TGCN'").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[2,1],grid=False,widths=0.5)
ax[2,1].set_title('TGCN')
df.query("dataset=='wikimath' and mtype=='rand' and inter_method == 'linear' and nof_filters==12 and lags==8 and epoch==50 and model=='DCRNN'").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax[2,2],grid=False,widths=0.5)
ax[2,2].set_title('DCRNN')
for ax in ax.flat:
ax.set_yticklabels([])
ax.axvline(x=2.5, color='black', linestyle='-')
ax.axvline(x=4.5, color='black', linestyle='-')
ax.axvline(x=6.5, color='black', linestyle='-')
ax.axvline(x=8.5, color='black', linestyle='-')
# ax.set_xticklabels(['IT-TGNN','TGNN','IT-TGNN','TGNN','IT-TGNN','TGNN','IT-TGNN','TGNN','IT-TGNN','TGNN'])
ax.set_xlabel('')
ax.set_ylabel('')
fig.suptitle('',fontsize=40)Text(0.5, 0.98, '')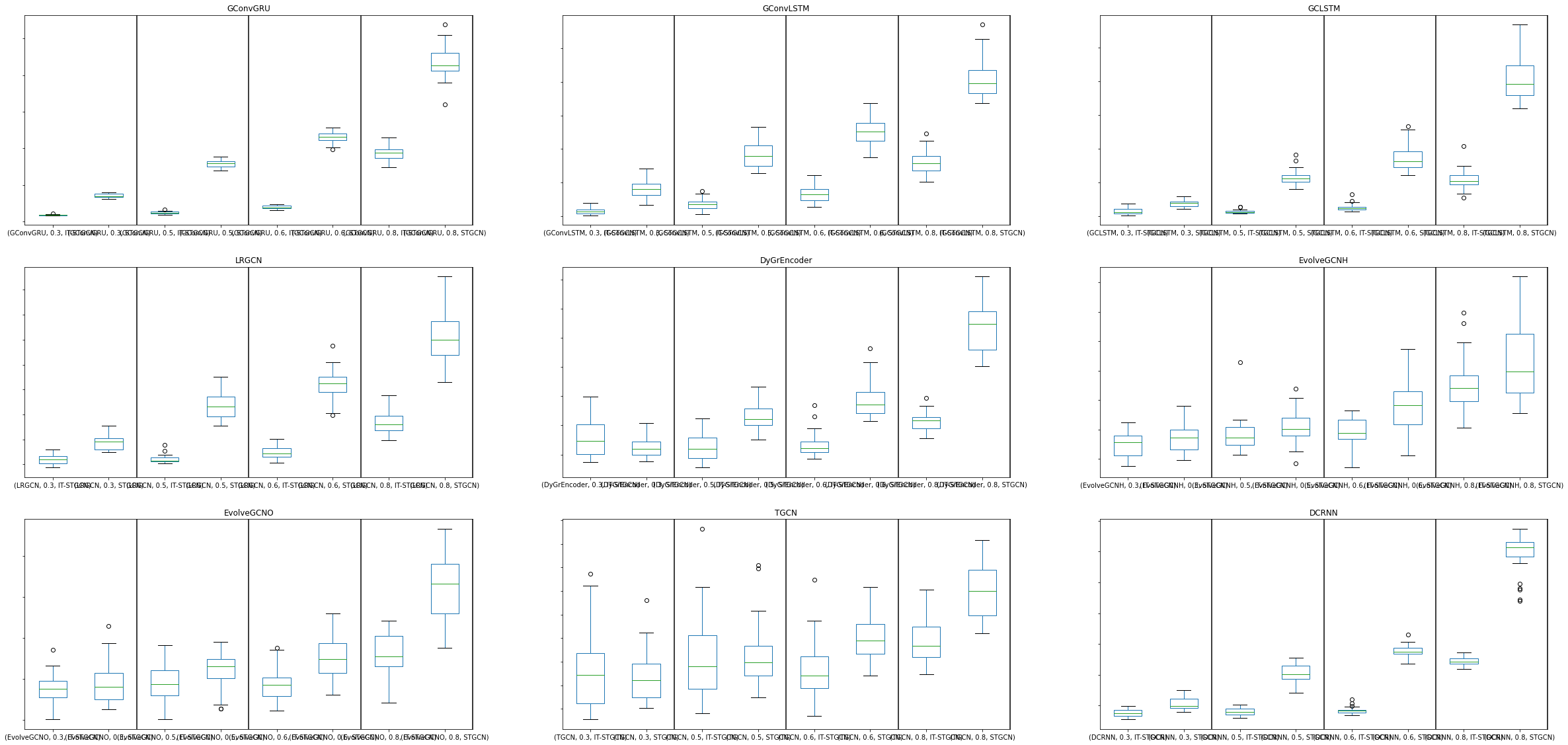
pd.merge(df.query("dataset=='wikimath' and mtype=='rand'").groupby(['model','mrate','lags','nof_filters','inter_method','method'])['mse'].mean().reset_index(),
df.query("dataset=='wikimath' and mtype=='rand'").groupby(['model','mrate','lags','nof_filters','inter_method','method'])['mse'].std().reset_index(),
on=['model','method','nof_filters','mrate','inter_method','lags']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3).query("mrate == 0.3")| model | mrate | lags | nof_filters | inter_method | method | mean | std | |
|---|---|---|---|---|---|---|---|---|
| 0 | DCRNN | 0.3 | 8 | 12.0 | linear | IT-STGCN | 0.588 | 0.007 |
| 1 | DCRNN | 0.3 | 8 | 12.0 | linear | STGCN | 0.603 | 0.010 |
| 8 | DyGrEncoder | 0.3 | 8 | 12.0 | linear | IT-STGCN | 0.578 | 0.031 |
| 9 | DyGrEncoder | 0.3 | 8 | 12.0 | linear | STGCN | 0.562 | 0.016 |
| 16 | EvolveGCNH | 0.3 | 8 | 12.0 | linear | IT-STGCN | 0.775 | 0.021 |
| 17 | EvolveGCNH | 0.3 | 8 | 12.0 | linear | STGCN | 0.787 | 0.024 |
| 24 | EvolveGCNO | 0.3 | 8 | 12.0 | linear | IT-STGCN | 0.738 | 0.018 |
| 25 | EvolveGCNO | 0.3 | 8 | 12.0 | linear | STGCN | 0.743 | 0.024 |
| 32 | GCLSTM | 0.3 | 8 | 64.0 | linear | IT-STGCN | 0.628 | 0.020 |
| 33 | GCLSTM | 0.3 | 8 | 64.0 | linear | STGCN | 0.674 | 0.020 |
| 40 | GConvGRU | 0.3 | 8 | 12.0 | linear | IT-STGCN | 0.518 | 0.002 |
| 41 | GConvGRU | 0.3 | 8 | 12.0 | linear | STGCN | 0.570 | 0.006 |
| 48 | GConvLSTM | 0.3 | 8 | 64.0 | linear | IT-STGCN | 0.631 | 0.019 |
| 49 | GConvLSTM | 0.3 | 8 | 64.0 | linear | STGCN | 0.764 | 0.057 |
| 56 | LRGCN | 0.3 | 8 | 32.0 | linear | IT-STGCN | 0.619 | 0.019 |
| 57 | LRGCN | 0.3 | 8 | 32.0 | linear | STGCN | 0.689 | 0.032 |
| 64 | TGCN | 0.3 | 8 | 12.0 | linear | IT-STGCN | 0.739 | 0.040 |
| 65 | TGCN | 0.3 | 8 | 12.0 | linear | STGCN | 0.734 | 0.027 |
pd.merge(df.query("dataset=='wikimath' and mtype=='rand'").groupby(['model','mrate','lags','inter_method','method'])['mse'].mean().reset_index(),
df.query("dataset=='wikimath' and mtype=='rand'").groupby(['model','mrate','lags','inter_method','method'])['mse'].std().reset_index(),
on=['model','method','mrate','inter_method','lags']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3).query("mrate != 0.3")| model | mrate | lags | inter_method | method | mean | std | |
|---|---|---|---|---|---|---|---|
| 2 | DCRNN | 0.5 | 8 | linear | IT-STGCN | 0.590 | 0.006 |
| 3 | DCRNN | 0.5 | 8 | linear | STGCN | 0.652 | 0.015 |
| 4 | DCRNN | 0.6 | 8 | linear | IT-STGCN | 0.592 | 0.005 |
| 5 | DCRNN | 0.6 | 8 | linear | STGCN | 0.688 | 0.011 |
| 6 | DCRNN | 0.8 | 8 | linear | IT-STGCN | 0.672 | 0.007 |
| 7 | DCRNN | 0.8 | 8 | linear | STGCN | 0.846 | 0.031 |
| 10 | DyGrEncoder | 0.5 | 8 | linear | IT-STGCN | 0.565 | 0.024 |
| 11 | DyGrEncoder | 0.5 | 8 | linear | STGCN | 0.614 | 0.024 |
| 12 | DyGrEncoder | 0.6 | 8 | linear | IT-STGCN | 0.566 | 0.021 |
| 13 | DyGrEncoder | 0.6 | 8 | linear | STGCN | 0.644 | 0.032 |
| 14 | DyGrEncoder | 0.8 | 8 | linear | IT-STGCN | 0.606 | 0.017 |
| 15 | DyGrEncoder | 0.8 | 8 | linear | STGCN | 0.770 | 0.045 |
| 18 | EvolveGCNH | 0.5 | 8 | linear | IT-STGCN | 0.791 | 0.029 |
| 19 | EvolveGCNH | 0.5 | 8 | linear | STGCN | 0.804 | 0.026 |
| 20 | EvolveGCNH | 0.6 | 8 | linear | IT-STGCN | 0.795 | 0.025 |
| 21 | EvolveGCNH | 0.6 | 8 | linear | STGCN | 0.843 | 0.042 |
| 22 | EvolveGCNH | 0.8 | 8 | linear | IT-STGCN | 0.877 | 0.045 |
| 23 | EvolveGCNH | 0.8 | 8 | linear | STGCN | 0.915 | 0.063 |
| 26 | EvolveGCNO | 0.5 | 8 | linear | IT-STGCN | 0.744 | 0.021 |
| 27 | EvolveGCNO | 0.5 | 8 | linear | STGCN | 0.759 | 0.021 |
| 28 | EvolveGCNO | 0.6 | 8 | linear | IT-STGCN | 0.745 | 0.019 |
| 29 | EvolveGCNO | 0.6 | 8 | linear | STGCN | 0.775 | 0.026 |
| 30 | EvolveGCNO | 0.8 | 8 | linear | IT-STGCN | 0.780 | 0.027 |
| 31 | EvolveGCNO | 0.8 | 8 | linear | STGCN | 0.863 | 0.038 |
| 34 | GCLSTM | 0.5 | 8 | linear | IT-STGCN | 0.626 | 0.010 |
| 35 | GCLSTM | 0.5 | 8 | linear | STGCN | 0.825 | 0.045 |
| 36 | GCLSTM | 0.6 | 8 | linear | IT-STGCN | 0.650 | 0.021 |
| 37 | GCLSTM | 0.6 | 8 | linear | STGCN | 0.942 | 0.078 |
| 38 | GCLSTM | 0.8 | 8 | linear | IT-STGCN | 0.815 | 0.058 |
| 39 | GCLSTM | 0.8 | 8 | linear | STGCN | 1.407 | 0.117 |
| 42 | GConvGRU | 0.5 | 8 | linear | IT-STGCN | 0.524 | 0.003 |
| 43 | GConvGRU | 0.5 | 8 | linear | STGCN | 0.658 | 0.010 |
| 44 | GConvGRU | 0.6 | 8 | linear | IT-STGCN | 0.539 | 0.004 |
| 45 | GConvGRU | 0.6 | 8 | linear | STGCN | 0.731 | 0.015 |
| 46 | GConvGRU | 0.8 | 8 | linear | IT-STGCN | 0.687 | 0.021 |
| 47 | GConvGRU | 0.8 | 8 | linear | STGCN | 0.932 | 0.043 |
| 50 | GConvLSTM | 0.5 | 8 | linear | IT-STGCN | 0.669 | 0.034 |
| 51 | GConvLSTM | 0.5 | 8 | linear | STGCN | 0.965 | 0.077 |
| 52 | GConvLSTM | 0.6 | 8 | linear | IT-STGCN | 0.731 | 0.048 |
| 53 | GConvLSTM | 0.6 | 8 | linear | STGCN | 1.104 | 0.078 |
| 54 | GConvLSTM | 0.8 | 8 | linear | IT-STGCN | 0.920 | 0.069 |
| 55 | GConvLSTM | 0.8 | 8 | linear | STGCN | 1.423 | 0.121 |
| 57 | GNAR | 0.8 | 8 | linear | GNAR | 1.354 | 0.000 |
| 60 | LRGCN | 0.5 | 8 | linear | IT-STGCN | 0.621 | 0.015 |
| 61 | LRGCN | 0.5 | 8 | linear | STGCN | 0.838 | 0.054 |
| 62 | LRGCN | 0.6 | 8 | linear | IT-STGCN | 0.648 | 0.024 |
| 63 | LRGCN | 0.6 | 8 | linear | STGCN | 0.917 | 0.063 |
| 64 | LRGCN | 0.8 | 8 | linear | IT-STGCN | 0.769 | 0.045 |
| 65 | LRGCN | 0.8 | 8 | linear | STGCN | 1.105 | 0.099 |
| 68 | TGCN | 0.5 | 8 | linear | IT-STGCN | 0.757 | 0.046 |
| 69 | TGCN | 0.5 | 8 | linear | STGCN | 0.757 | 0.034 |
| 70 | TGCN | 0.6 | 8 | linear | IT-STGCN | 0.742 | 0.030 |
| 71 | TGCN | 0.6 | 8 | linear | STGCN | 0.774 | 0.025 |
| 72 | TGCN | 0.8 | 8 | linear | IT-STGCN | 0.771 | 0.020 |
| 73 | TGCN | 0.8 | 8 | linear | STGCN | 0.827 | 0.030 |
Block
pd.merge(df.query("dataset=='wikimath' and mtype=='block'").groupby(['model','mrate','lags','inter_method','method'])['mse'].mean().reset_index(),
df.query("dataset=='wikimath' and mtype=='block'").groupby(['model','mrate','lags','inter_method','method'])['mse'].std().reset_index(),
on=['model','method','mrate','inter_method','lags']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3)| model | mrate | lags | inter_method | method | mean | std | |
|---|---|---|---|---|---|---|---|
| 0 | DCRNN | 0.120 | 8 | linear | IT-STGCN | 0.583 | 0.006 |
| 1 | DCRNN | 0.120 | 8 | linear | STGCN | 0.578 | 0.005 |
| 2 | DyGrEncoder | 0.120 | 8 | linear | IT-STGCN | 0.563 | 0.025 |
| 3 | DyGrEncoder | 0.120 | 8 | linear | STGCN | 0.546 | 0.016 |
| 4 | EvolveGCNH | 0.120 | 8 | linear | IT-STGCN | 0.776 | 0.028 |
| 5 | EvolveGCNH | 0.120 | 8 | linear | STGCN | 0.773 | 0.021 |
| 6 | EvolveGCNO | 0.120 | 8 | linear | IT-STGCN | 0.732 | 0.025 |
| 7 | EvolveGCNO | 0.120 | 8 | linear | STGCN | 0.735 | 0.022 |
| 8 | GCLSTM | 0.120 | 8 | linear | IT-STGCN | 0.640 | 0.019 |
| 9 | GCLSTM | 0.120 | 8 | linear | STGCN | 0.638 | 0.013 |
| 10 | GConvGRU | 0.004 | 8 | linear | IT-STGCN | 0.529 | 0.003 |
| 11 | GConvGRU | 0.004 | 8 | linear | STGCN | 0.528 | 0.003 |
| 12 | GConvGRU | 0.096 | 8 | linear | IT-STGCN | 0.529 | 0.004 |
| 13 | GConvGRU | 0.096 | 8 | linear | STGCN | 0.544 | 0.011 |
| 14 | GConvGRU | 0.120 | 8 | linear | IT-STGCN | 0.523 | 0.002 |
| 15 | GConvGRU | 0.120 | 8 | linear | STGCN | 0.531 | 0.002 |
| 16 | GConvLSTM | 0.120 | 8 | linear | IT-STGCN | 0.627 | 0.014 |
| 17 | GConvLSTM | 0.120 | 8 | linear | STGCN | 0.660 | 0.034 |
| 18 | GNAR | 0.120 | 8 | linear | GNAR | 1.354 | 0.000 |
| 19 | LRGCN | 0.120 | 8 | linear | IT-STGCN | 0.608 | 0.012 |
| 20 | LRGCN | 0.120 | 8 | linear | STGCN | 0.624 | 0.024 |
| 21 | TGCN | 0.120 | 8 | linear | IT-STGCN | 0.748 | 0.046 |
| 22 | TGCN | 0.120 | 8 | linear | STGCN | 0.741 | 0.046 |
missing values on the same nodes
pd.merge(df2.query("dataset=='wikimath'").groupby(['model','mrate','lags','method'])['mse'].mean().reset_index(),
df2.query("dataset=='wikimath'").groupby(['model','mrate','lags','method'])['mse'].std().reset_index(),
on=['model','method','mrate','lags']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3)| model | mrate | lags | method | mean | std | |
|---|---|---|---|---|---|---|
| 0 | DCRNN | 0.512 | 8 | IT-STGCN | 0.592 | 0.005 |
| 1 | DCRNN | 0.512 | 8 | STGCN | 0.665 | 0.015 |
| 2 | DyGrEncoder | 0.512 | 8 | IT-STGCN | 0.561 | 0.031 |
| 3 | DyGrEncoder | 0.512 | 8 | STGCN | 0.626 | 0.027 |
| 4 | EvolveGCNH | 0.512 | 8 | IT-STGCN | 0.794 | 0.031 |
| 5 | EvolveGCNH | 0.512 | 8 | STGCN | 0.818 | 0.031 |
| 6 | EvolveGCNO | 0.512 | 8 | IT-STGCN | 0.745 | 0.017 |
| 7 | EvolveGCNO | 0.512 | 8 | STGCN | 0.753 | 0.026 |
| 8 | GCLSTM | 0.512 | 8 | IT-STGCN | 0.617 | 0.011 |
| 9 | GCLSTM | 0.512 | 8 | STGCN | 0.823 | 0.048 |
| 10 | GConvGRU | 0.512 | 8 | IT-STGCN | 0.533 | 0.003 |
| 11 | GConvGRU | 0.512 | 8 | STGCN | 0.726 | 0.015 |
| 12 | GConvLSTM | 0.512 | 8 | IT-STGCN | 0.653 | 0.033 |
| 13 | GConvLSTM | 0.512 | 8 | STGCN | 0.963 | 0.098 |
| 14 | GNAR | 0.512 | 8 | GNAR | 1.354 | 0.000 |
| 15 | LRGCN | 0.512 | 8 | IT-STGCN | 0.624 | 0.019 |
| 16 | LRGCN | 0.512 | 8 | STGCN | 0.810 | 0.064 |
| 17 | TGCN | 0.512 | 8 | IT-STGCN | 0.750 | 0.039 |
| 18 | TGCN | 0.512 | 8 | STGCN | 0.782 | 0.030 |
Windmillsmall(lags=8)
df.query("model=='GNAR' and dataset=='windmillsmall'")| dataset | method | mrate | mtype | lags | nof_filters | inter_method | epoch | mse | calculation_time | model | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 26584 | windmillsmall | GNAR | 0.081291 | block | 8 | NaN | linear | NaN | 1.64923 | 3.428970 | GNAR |
| 26585 | windmillsmall | GNAR | 0.081291 | block | 8 | NaN | linear | NaN | 1.64923 | 3.492012 | GNAR |
| 26586 | windmillsmall | GNAR | 0.081291 | block | 8 | NaN | linear | NaN | 1.64923 | 2.916400 | GNAR |
| 26587 | windmillsmall | GNAR | 0.000000 | NaN | 8 | NaN | NaN | NaN | 1.64923 | 2.513212 | GNAR |
| 26588 | windmillsmall | GNAR | 0.700000 | rand | 8 | NaN | nearest | NaN | 1.64923 | 3.334994 | GNAR |
| 26589 | windmillsmall | GNAR | 0.000000 | NaN | 8 | NaN | NaN | NaN | 1.64923 | 2.116200 | GNAR |
| 26590 | windmillsmall | GNAR | 0.700000 | rand | 8 | NaN | nearest | NaN | 1.64923 | 2.391699 | GNAR |
| 26591 | windmillsmall | GNAR | 0.000000 | NaN | 8 | NaN | NaN | NaN | 1.64923 | 3.406726 | GNAR |
| 26592 | windmillsmall | GNAR | 0.700000 | rand | 8 | NaN | nearest | NaN | 1.64923 | 2.586957 | GNAR |
Baseline
pd.merge(df.query("dataset=='windmillsmall' and mrate==0").groupby(['model','lags'])['mse'].mean().reset_index(),
df.query("dataset=='windmillsmall' and mrate==0").groupby(['model','lags'])['mse'].std().reset_index(),
on=['model','lags']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3)| model | lags | mean | std | |
|---|---|---|---|---|
| 0 | DCRNN | 8 | 0.988 | 0.003 |
| 1 | DyGrEncoder | 8 | 0.988 | 0.008 |
| 2 | EvolveGCNH | 8 | 0.986 | 0.002 |
| 3 | EvolveGCNO | 8 | 0.983 | 0.001 |
| 4 | GCLSTM | 8 | 0.992 | 0.010 |
| 5 | GConvGRU | 8 | 1.003 | 0.004 |
| 6 | GConvLSTM | 8 | 1.019 | 0.045 |
| 7 | GNAR | 8 | 1.649 | 0.000 |
| 8 | LRGCN | 8 | 0.987 | 0.006 |
| 9 | TGCN | 8 | 0.991 | 0.010 |
Random
fig, ((ax1)) = plt.subplots(nrows=1, ncols=1, figsize=(50, 6), sharey=True)
df.query("dataset=='windmillsmall' and mtype=='rand' and inter_method == 'linear' and lags==8 and epoch==50").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax1,grid=False,widths=0.5)
pd.merge(df.query("dataset=='windmillsmall' and mtype=='rand'").groupby(['model','mrate','lags','inter_method','method'])['mse'].mean().reset_index(),
df.query("dataset=='windmillsmall' and mtype=='rand'").groupby(['model','mrate','lags','inter_method','method'])['mse'].std().reset_index(),
on=['model','method','mrate','inter_method','lags']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3)| model | mrate | lags | inter_method | method | mean | std | |
|---|---|---|---|---|---|---|---|
| 0 | DCRNN | 0.7 | 8 | linear | IT-STGCN | 1.117 | 0.034 |
| 1 | DCRNN | 0.7 | 8 | linear | STGCN | 1.348 | 0.057 |
| 2 | DyGrEncoder | 0.7 | 8 | linear | IT-STGCN | 1.127 | 0.009 |
| 3 | DyGrEncoder | 0.7 | 8 | linear | STGCN | 1.709 | 0.074 |
| 4 | EvolveGCNH | 0.7 | 8 | linear | IT-STGCN | 1.129 | 0.035 |
| 5 | EvolveGCNH | 0.7 | 8 | linear | STGCN | 1.330 | 0.137 |
| 6 | EvolveGCNO | 0.7 | 8 | linear | IT-STGCN | 1.149 | 0.026 |
| 7 | EvolveGCNO | 0.7 | 8 | linear | STGCN | 1.495 | 0.137 |
| 8 | GCLSTM | 0.7 | 8 | linear | IT-STGCN | 1.116 | 0.021 |
| 9 | GCLSTM | 0.7 | 8 | linear | STGCN | 1.573 | 0.105 |
| 10 | GConvGRU | 0.7 | 8 | linear | IT-STGCN | 1.194 | 0.042 |
| 11 | GConvGRU | 0.7 | 8 | linear | STGCN | 1.662 | 0.073 |
| 12 | GConvLSTM | 0.7 | 8 | linear | IT-STGCN | 1.142 | 0.021 |
| 13 | GConvLSTM | 0.7 | 8 | linear | STGCN | 1.599 | 0.057 |
| 14 | GNAR | 0.7 | 8 | nearest | GNAR | 1.649 | 0.000 |
| 15 | LRGCN | 0.7 | 8 | linear | IT-STGCN | 1.110 | 0.012 |
| 16 | LRGCN | 0.7 | 8 | linear | STGCN | 1.492 | 0.087 |
| 17 | TGCN | 0.7 | 8 | linear | IT-STGCN | 1.071 | 0.010 |
| 18 | TGCN | 0.7 | 8 | linear | STGCN | 1.305 | 0.039 |
Block
pd.merge(df.query("dataset=='windmillsmall' and mtype=='block'").groupby(['model','mrate','nof_filters','lags','method'])['mse'].mean().reset_index(),
df.query("dataset=='windmillsmall' and mtype=='block'").groupby(['model','mrate','nof_filters','lags','method'])['mse'].std().reset_index(),
on=['model','method','nof_filters','mrate','lags']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3)| model | mrate | nof_filters | lags | method | mean | std | |
|---|---|---|---|---|---|---|---|
| 0 | DCRNN | 0.081 | 4.0 | 8 | IT-STGCN | 0.983 | 0.002 |
| 1 | DCRNN | 0.081 | 4.0 | 8 | STGCN | 0.994 | 0.005 |
| 2 | DyGrEncoder | 0.081 | 12.0 | 8 | IT-STGCN | 0.985 | 0.005 |
| 3 | DyGrEncoder | 0.081 | 12.0 | 8 | STGCN | 0.985 | 0.003 |
| 4 | EvolveGCNH | 0.081 | 12.0 | 8 | IT-STGCN | 0.986 | 0.003 |
| 5 | EvolveGCNH | 0.081 | 12.0 | 8 | STGCN | 0.993 | 0.003 |
| 6 | EvolveGCNO | 0.081 | 12.0 | 8 | IT-STGCN | 0.983 | 0.002 |
| 7 | EvolveGCNO | 0.081 | 12.0 | 8 | STGCN | 0.990 | 0.002 |
| 8 | GCLSTM | 0.081 | 16.0 | 8 | IT-STGCN | 0.985 | 0.003 |
| 9 | GCLSTM | 0.081 | 16.0 | 8 | STGCN | 0.985 | 0.002 |
| 10 | GConvGRU | 0.081 | 12.0 | 8 | IT-STGCN | 1.007 | 0.005 |
| 11 | GConvGRU | 0.081 | 12.0 | 8 | STGCN | 1.008 | 0.006 |
| 12 | GConvLSTM | 0.081 | 16.0 | 8 | IT-STGCN | 0.997 | 0.022 |
| 13 | GConvLSTM | 0.081 | 16.0 | 8 | STGCN | 0.989 | 0.009 |
| 14 | LRGCN | 0.081 | 12.0 | 8 | IT-STGCN | 0.985 | 0.002 |
| 15 | LRGCN | 0.081 | 12.0 | 8 | STGCN | 0.985 | 0.003 |
| 16 | TGCN | 0.081 | 12.0 | 8 | IT-STGCN | 0.992 | 0.015 |
| 17 | TGCN | 0.081 | 12.0 | 8 | STGCN | 0.999 | 0.013 |
Montevideobus (lags=4)
df.query("model=='GNAR' and dataset=='monte'")| dataset | method | mrate | mtype | lags | nof_filters | inter_method | epoch | mse | calculation_time | model | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 26554 | monte | GNAR | 0.000000 | NaN | 4 | NaN | NaN | NaN | 1.061937 | 6.576898 | GNAR |
| 26555 | monte | GNAR | 0.800000 | rand | 4 | NaN | nearest | NaN | 1.061937 | 4.220497 | GNAR |
| 26556 | monte | GNAR | 0.000000 | NaN | 4 | NaN | NaN | NaN | 1.061937 | 4.429234 | GNAR |
| 26557 | monte | GNAR | 0.800000 | rand | 4 | NaN | nearest | NaN | 1.061937 | 4.886660 | GNAR |
| 26558 | monte | GNAR | 0.000000 | NaN | 4 | NaN | NaN | NaN | 1.061937 | 4.879432 | GNAR |
| 26559 | monte | GNAR | 0.800000 | rand | 4 | NaN | nearest | NaN | 1.061937 | 4.814065 | GNAR |
| 26581 | monte | GNAR | 0.149142 | block | 4 | NaN | nearest | NaN | 1.061937 | 4.948565 | GNAR |
| 26582 | monte | GNAR | 0.149142 | block | 4 | NaN | nearest | NaN | 1.061937 | 2.942073 | GNAR |
| 26583 | monte | GNAR | 0.149142 | block | 4 | NaN | nearest | NaN | 1.061937 | 5.179985 | GNAR |
Baseline
pd.merge(df.query("dataset=='monte' and mrate==0").groupby(['model','lags'])['mse'].mean().reset_index(),
df.query("dataset=='monte' and mrate==0").groupby(['model','lags'])['mse'].std().reset_index(),
on=['model','lags']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3)| model | lags | mean | std | |
|---|---|---|---|---|
| 0 | DCRNN | 4 | 0.936 | 0.002 |
| 1 | DyGrEncoder | 4 | 0.995 | 0.034 |
| 2 | EvolveGCNH | 4 | 1.182 | 0.192 |
| 3 | EvolveGCNO | 4 | 1.157 | 0.182 |
| 4 | GCLSTM | 4 | 0.970 | 0.011 |
| 5 | GConvGRU | 4 | 0.931 | 0.002 |
| 6 | GConvLSTM | 4 | 0.960 | 0.011 |
| 7 | GNAR | 4 | 1.062 | 0.000 |
| 8 | LRGCN | 4 | 0.980 | 0.024 |
| 9 | TGCN | 4 | 0.983 | 0.006 |
Random
fig, ((ax1)) = plt.subplots(nrows=1, ncols=1, figsize=(50, 6), sharey=True)
df.query("dataset=='monte' and mtype=='rand' and inter_method == 'nearest' and lags==4 and epoch==50").\
iloc[:,[1,2,8,10]].boxplot(by=['model','mrate','method'],ax=ax1,grid=False,widths=0.5)
pd.merge(df.query("dataset=='monte' and mtype=='rand'").groupby(['model','mrate','nof_filters','lags','inter_method','method'])['mse'].mean().reset_index(),
df.query("dataset=='monte' and mtype=='rand'").groupby(['model','mrate','nof_filters','lags','inter_method','method'])['mse'].std().reset_index(),
on=['model','mrate','nof_filters','inter_method','method','mrate','lags']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3)| model | mrate | nof_filters | lags | inter_method | method | mean | std | |
|---|---|---|---|---|---|---|---|---|
| 0 | DCRNN | 0.3 | 12.0 | 4 | nearest | IT-STGCN | 0.938 | 0.001 |
| 1 | DCRNN | 0.3 | 12.0 | 4 | nearest | STGCN | 1.008 | 0.006 |
| 2 | DCRNN | 0.5 | 12.0 | 4 | nearest | IT-STGCN | 0.948 | 0.002 |
| 3 | DCRNN | 0.5 | 12.0 | 4 | nearest | STGCN | 1.119 | 0.025 |
| 4 | DCRNN | 0.7 | 12.0 | 4 | nearest | IT-STGCN | 1.031 | 0.018 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 67 | TGCN | 0.5 | 8.0 | 4 | nearest | STGCN | 1.100 | 0.036 |
| 68 | TGCN | 0.7 | 8.0 | 4 | nearest | IT-STGCN | 1.030 | 0.010 |
| 69 | TGCN | 0.7 | 8.0 | 4 | nearest | STGCN | 1.183 | 0.057 |
| 70 | TGCN | 0.8 | 8.0 | 4 | nearest | IT-STGCN | 1.073 | 0.024 |
| 71 | TGCN | 0.8 | 8.0 | 4 | nearest | STGCN | 1.218 | 0.086 |
72 rows × 8 columns
Block
pd.merge(df.query("dataset=='monte' and mtype=='block'").groupby(['model','mrate','lags','inter_method','method'])['mse'].mean().reset_index(),
df.query("dataset=='monte' and mtype=='block'").groupby(['model','mrate','lags','inter_method','method'])['mse'].std().reset_index(),
on=['model','method','mrate','inter_method','lags']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3)| model | mrate | lags | inter_method | method | mean | std | |
|---|---|---|---|---|---|---|---|
| 0 | DCRNN | 0.149 | 4 | nearest | IT-STGCN | 0.940 | 0.001 |
| 1 | DCRNN | 0.149 | 4 | nearest | STGCN | 0.956 | 0.003 |
| 2 | DyGrEncoder | 0.149 | 4 | nearest | IT-STGCN | 1.005 | 0.046 |
| 3 | DyGrEncoder | 0.149 | 4 | nearest | STGCN | 1.030 | 0.044 |
| 4 | EvolveGCNH | 0.149 | 4 | nearest | IT-STGCN | 1.392 | 0.110 |
| 5 | EvolveGCNH | 0.149 | 4 | nearest | STGCN | 1.612 | 0.216 |
| 6 | EvolveGCNO | 0.149 | 4 | nearest | IT-STGCN | 1.345 | 0.110 |
| 7 | EvolveGCNO | 0.149 | 4 | nearest | STGCN | 1.766 | 0.123 |
| 8 | GCLSTM | 0.149 | 4 | nearest | IT-STGCN | 0.959 | 0.008 |
| 9 | GCLSTM | 0.149 | 4 | nearest | STGCN | 0.956 | 0.005 |
| 10 | GConvGRU | 0.149 | 4 | cubic | IT-STGCN | 1.023 | 0.021 |
| 11 | GConvGRU | 0.149 | 4 | cubic | STGCN | 1.028 | 0.031 |
| 12 | GConvGRU | 0.149 | 4 | linear | IT-STGCN | 0.930 | 0.002 |
| 13 | GConvGRU | 0.149 | 4 | linear | STGCN | 0.935 | 0.005 |
| 14 | GConvGRU | 0.149 | 4 | nearest | IT-STGCN | 0.932 | 0.002 |
| 15 | GConvGRU | 0.149 | 4 | nearest | STGCN | 0.935 | 0.004 |
| 16 | GConvLSTM | 0.149 | 4 | nearest | IT-STGCN | 0.949 | 0.008 |
| 17 | GConvLSTM | 0.149 | 4 | nearest | STGCN | 0.950 | 0.005 |
| 18 | GNAR | 0.149 | 4 | nearest | GNAR | 1.062 | 0.000 |
| 19 | LRGCN | 0.149 | 4 | nearest | IT-STGCN | 0.978 | 0.024 |
| 20 | LRGCN | 0.149 | 4 | nearest | STGCN | 0.977 | 0.020 |
| 21 | TGCN | 0.149 | 4 | nearest | IT-STGCN | 0.984 | 0.007 |
| 22 | TGCN | 0.149 | 4 | nearest | STGCN | 0.985 | 0.005 |
pd.merge(df.query("dataset=='monte' and mtype=='block'").groupby(['model','mrate','lags','inter_method','method'])['mse'].mean().reset_index(),
df.query("dataset=='monte' and mtype=='block'").groupby(['model','mrate','lags','inter_method','method'])['mse'].std().reset_index(),
on=['model','method','mrate','inter_method','lags']).rename(columns={'mse_x':'mean','mse_y':'std'}).round(3).query("inter_method=='nearest'")| model | mrate | lags | inter_method | method | mean | std | |
|---|---|---|---|---|---|---|---|
| 0 | DCRNN | 0.149 | 4 | nearest | IT-STGCN | 0.940 | 0.001 |
| 1 | DCRNN | 0.149 | 4 | nearest | STGCN | 0.956 | 0.003 |
| 2 | DyGrEncoder | 0.149 | 4 | nearest | IT-STGCN | 1.005 | 0.046 |
| 3 | DyGrEncoder | 0.149 | 4 | nearest | STGCN | 1.030 | 0.044 |
| 4 | EvolveGCNH | 0.149 | 4 | nearest | IT-STGCN | 1.392 | 0.110 |
| 5 | EvolveGCNH | 0.149 | 4 | nearest | STGCN | 1.612 | 0.216 |
| 6 | EvolveGCNO | 0.149 | 4 | nearest | IT-STGCN | 1.345 | 0.110 |
| 7 | EvolveGCNO | 0.149 | 4 | nearest | STGCN | 1.766 | 0.123 |
| 8 | GCLSTM | 0.149 | 4 | nearest | IT-STGCN | 0.959 | 0.008 |
| 9 | GCLSTM | 0.149 | 4 | nearest | STGCN | 0.956 | 0.005 |
| 14 | GConvGRU | 0.149 | 4 | nearest | IT-STGCN | 0.932 | 0.002 |
| 15 | GConvGRU | 0.149 | 4 | nearest | STGCN | 0.935 | 0.004 |
| 16 | GConvLSTM | 0.149 | 4 | nearest | IT-STGCN | 0.949 | 0.008 |
| 17 | GConvLSTM | 0.149 | 4 | nearest | STGCN | 0.950 | 0.005 |
| 18 | GNAR | 0.149 | 4 | nearest | GNAR | 1.062 | 0.000 |
| 19 | LRGCN | 0.149 | 4 | nearest | IT-STGCN | 0.978 | 0.024 |
| 20 | LRGCN | 0.149 | 4 | nearest | STGCN | 0.977 | 0.020 |
| 21 | TGCN | 0.149 | 4 | nearest | IT-STGCN | 0.984 | 0.007 |
| 22 | TGCN | 0.149 | 4 | nearest | STGCN | 0.985 | 0.005 |